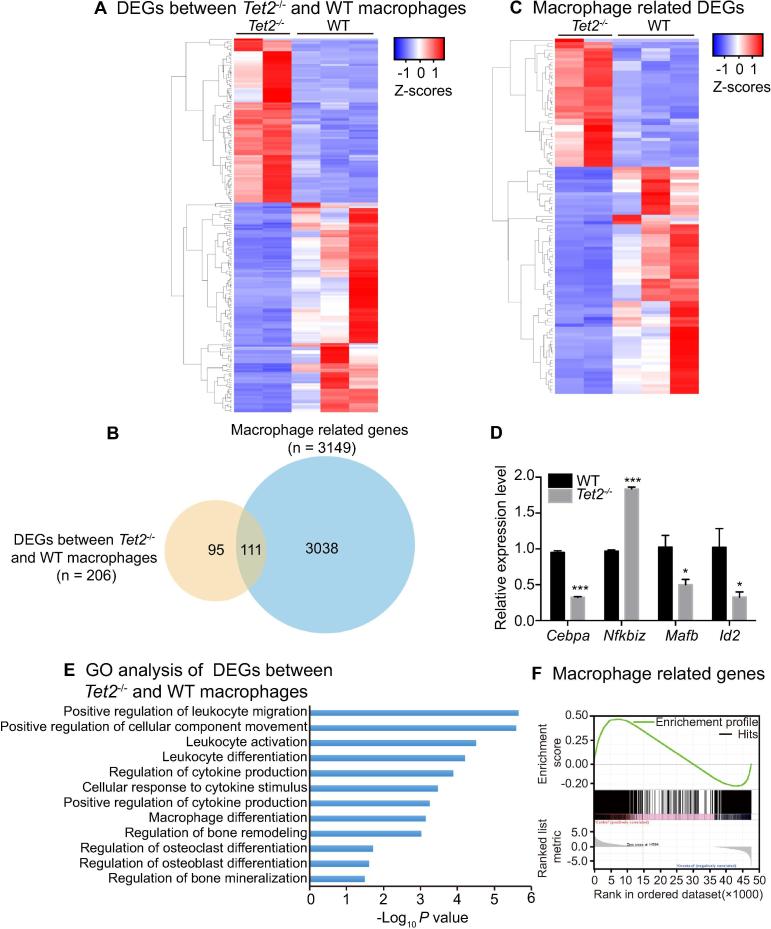
Figure 2.
Deletion of Tet2 alters gene expression profiling in macrophages
A. Heatmap showing the relative abundance of DEGs between WT and Tet2−/− macrophages by RNA-seq analysis (FDR < 0.05 across all three algorithms, CuffDiff, edgeR, and DESeq2). Z-scores per transcript are shown and blue represents lower expression and red represents higher expression. B. Venn diagram showing the overlap between the macrophage related genes and DEGs between Tet2−/− and WT macrophages. C. Heatmap showing the relative abundance of the 111 overlapping genes in panel B using RNA-seq analysis (FDR < 0.05 across all three algorithms, CuffDiff, edgeR, and DESeq2). D. Quantitative RT-PCR analysis of phenotypic associated DEGs (relative to Actb) in Tet2−/−vs. WT macrophages that are important for osteoclast differentiation. Data are presented as mean ± SEM, n = 3. *P < 0.05, ***P < 0.001. Student’s t test. E. Ingenuity Pathway Analysis (IPA) of gene ontology biological processes enriched in pathways that are involved in hematopoietic cell differentiation. F. GSEA analysis plot shows decreased gene expression of the macrophage related genes in macrophages from Tet2−/− mice compared to those from WT mice (NES = 1.4399285 and FDR < 1×10−4). DEG, differentially-expressed gene; GSEA, gene set enrichment analysis; NES, nominal enrichment score; FDR, false discovery rate.