Figure 5.
Tet2 associates with Runx1
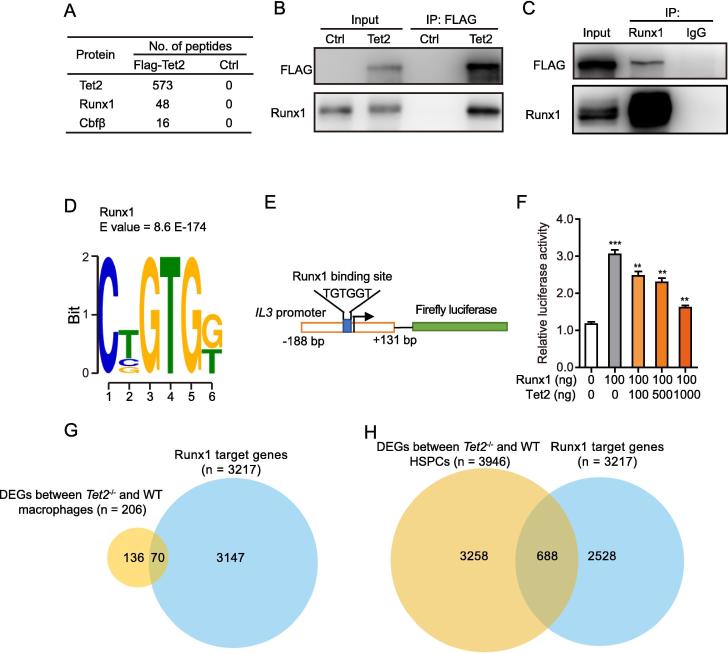
A. Proteins identified by mass spectrometry from MEL cells stably expressing FLAG-Tet2 after purification of Tet2-associated proteins. Spectral counts for each interacting protein are shown. B. and C. Reciprocal co-immunoprecipitations and Western blots confirming the interaction between Tet2 and Runx1 in HEK293T cells overexpressing FLAG-tagged Tet2. Flag (B) or Runx1 immunoprecipitation was performed for nuclear lysates of HEK293T cells overexpressing FLAG-vector (Ctrl) or FLAG-Tet2 (Tet2) using (C) before immunoblotting using the indicated antibodies. The isotype IgG was used as negative control in panel C. D. Motif enrichment analysis identifying the significant enrichment of Runx1 DNA binding motifs in the Tet2-binding sites. Tet2 ChIP-seq peaks were submitted to the MEME suite. E. Diagram of IL3-promoter-luciferase construct showing the Runx1 binding consensus site. F. HEK293T cells cotransfected with IL3-promoter luciferase reporter plasmid, 100 ng Runx1 and increasing concentrations of Tet2 expression plasmids. Renilla luciferase plasmid was used as an internal transfection control, and FC of reporter activity induced by Runx1/Tet2 expression relative to that of the Renilla luciferase control were plotted. Data are presented as mean ± SEM, n = 3. **P < 0.01, ***P < 0.001. G. Venn diagram showing the overlap between Runx1 target genes with DEGs between Tet2−/− and WT macrophages. H. Venn diagram showing the overlap between the Runx1 target genes with DEGs between Tet2−/− and WT HSPCs (lin−c-Kit+ gated). FC, fold change; HSPC, hematopoietic stem/progenitor cell.