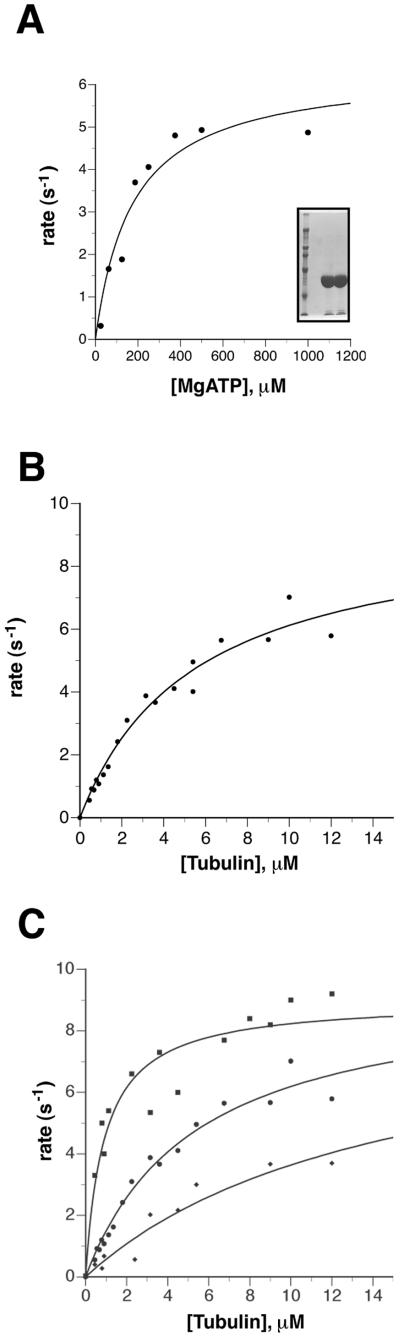
Figure 2.
Determination of the Km (ATP), Km (MT) for the minimal catalytic core of NOD (GST-NOD320): biochemical characterization of two NOD mutant proteins. (A) ATPase rate of 50 nM GST-NOD320 was measured in the presence of subsaturating levels of microtubules to determine the apparent Km for ATP. These values were plotted by DeltaGraph and Km values were obtained by curve fitting. The data was fit to the Michaelis-Menten kinetic model and the solid line was obtained from this curve-fitting procedure. The inset shows an SDS-PAGE gel showing molecular weight standards and the first two elutions of GST-NOD320. (B) ATPase rates of 50 nM GST-NOD320 were determined in the presence of various levels of polymerized tubulin and 0.5 mM ATP. The microtubule dimer concentration that led to half-maximum ATPase activation Km (MT) and the kcat were obtained by curve fitting with DeltaGraph. The solid line was obtained from the curve-fitting procedure (r = 0.97). (C) ATPase rates of various 50 nM GST-NOD fusions were measured and calculated as in B. Wild-type NOD-GST, NODDTW, and NOD“DR2” are represented by filled circles, diamonds, and squares, respectively. Km (MT) and the kcat were obtained by curve fitting with DeltaGraph with the following r values: GST-NOD320, 0.97; NODDTW, 0.92; and NOD“DR2”, 0.96.