Figure 1.
Workflow for the Selection of DARPins for Receptor-Targeted LVs and AAVs
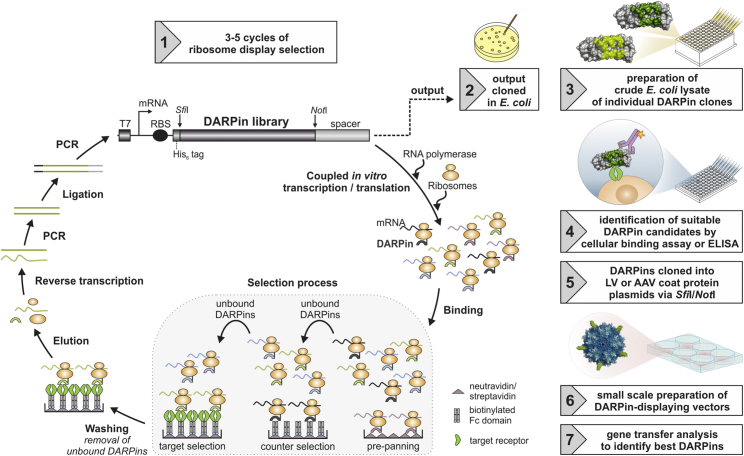
DARPin selection by ribosome display is shown in steps 1 and 2. All substeps of the ribosome display cycle are performed cell-free in vitro. Each cycle begins with the transcription and translation of a DARPin-encoding DNA library, flanked by a T7 promoter (T7), a ribosome binding site (RBS), and a spacer sequence. Ternary complexes of ribosome, the DARPin-encoding mRNA, and the translated polypeptide (shown in identical color) are formed and allowed to bind to the target protein during the selection process. The selection process encompasses three steps: pre-panning, counter-, and target selection. Pre-panning and counter-selection results in the elimination of the black and brown DARPins binding to the Fc domain or streptavidin used for immobilization of the target receptor. The green DARPin, in contrast, binds the target receptor and is carried forward to the next selection cycle. After the selection process, unbound complexes are washed away before elution, and reverse transcription of the mRNA is carried out. The cDNA fragments are PCR amplified and ligated to the upstream and downstream flanking sequences. The PCR-amplified ligation product is used as template library for the next ribosome display cycle or cloned into an expression vector for analysis of single clones on the protein level. After ribosome display, individual DARPin molecules are expressed as crude E. coli lysates (step 3), tested for their receptor binding ability (step 4), and subcloned into the corresponding viral vector plasmids (step 5) before small-scale generation of DARPin-displaying LV or AAV particles (step 6), which are finally analyzed for cell-type-specific gene transfer (step 7). Exemplarily an AAV vector is shown displaying five molecules of an individual DARPin clone on its surface, but the same procedure is applied for LV particles. Step 1 of the figure is adapted from Dreier and Plückthun.53