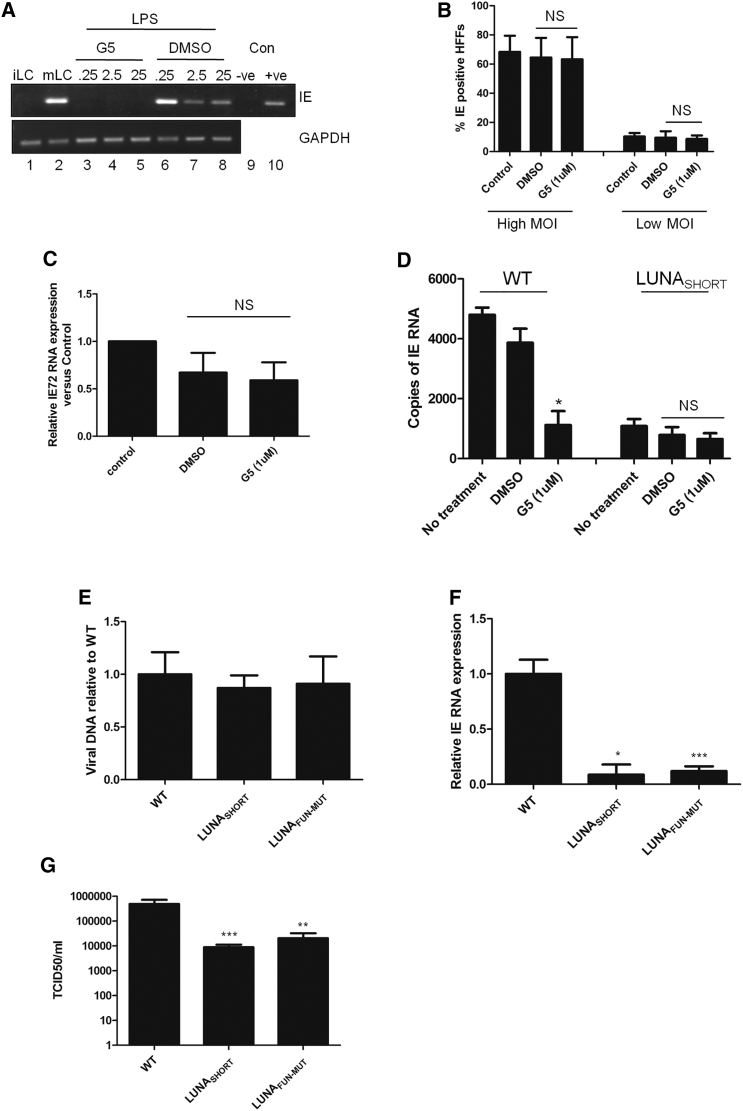
Figure 6.
LUNA Isopeptidase Activity Is Required for HCMV Reactivation
(A) CD34+ cells isolated from a healthy seropositive donor were cultured to immature DCs (iLC; lane 1) and then incubated with LPS to promote full virus reactivation (mLC; lanes 2–8). Prior to the addition of LPS, cells were incubated with mock (2), G5 isopeptidase inhibitor (25–0.25 μM; 3–5), or DMSO solvent (6–8) for 2 hr. Reactivation was measured using RT-PCR for IE72 expression on RNA isolated from cells 24 hr post-addition of LPS. Water (−ve) and cDNA from infected fibroblasts (+ve) served as PCR controls (lanes 9 and 10).
(B) HFFs were incubated with 1 μM G5 and infected with HCMV at an MOI of 1 (high MOI) or 0.1 (low MOI) and then analyzed by immunofluorescence (IF) at 8 hr post-infection for IE gene expression, and percentage infection was calculated.
(C) A qRT-PCR analysis for IE gene expression was performed on low-MOI infected HFF cells as described in (B).
(D) CD34+ cells latently infected with WT or the LUNA protein disruption virus (LUNASHORT) were differentiated to immature DCs and then either incubated with DMSO or G5 prior to stimulation with LPS to fully reactivate virus. Twenty-four hours post-infection RNA was isolated and analyzed using IE qRT-PCR and quantified against a standard curve. Data are mean ± SD and represent triplicate analyses performed in three independent experiments. ∗p < 0.05; NS, not significant.
(E–G) CD34+ cells from two donors were infected with Merlin, LUNASHORT, or LUNAFUN-MUT. Seven dpi cells were differentiated to immature DCs and analyzed using qPCR for viral genome carriage with viral genomes expressed relative to cellular DNA control (E). Alternatively, immature CD34+-derived DCs were incubated with LPS and then either analyzed for IE RNA expression by qRT-PCR (F) or co-cultured with HFFs and assayed for infectious virus production (G). Data are mean ± SD and represent triplicate analyses performed in two independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; NS, not significant (n = 2).
See also Figure S5.