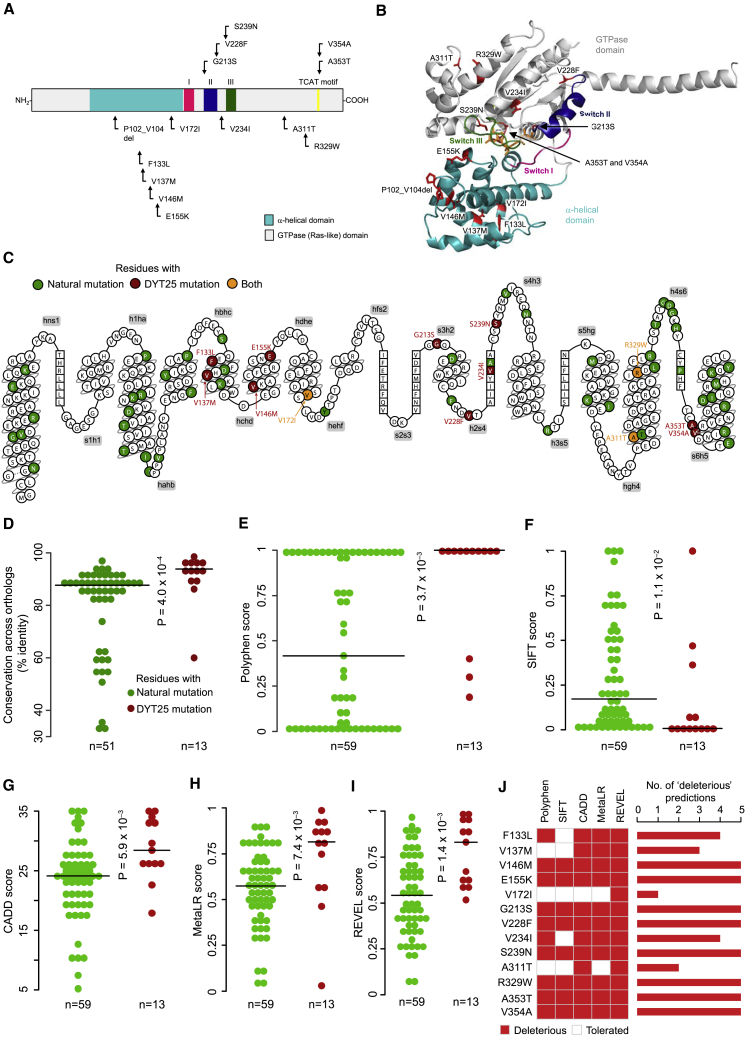
Figure 2.
Distribution of Gαolf Mutations and Computational Analysis of Naturally Occurring Mutations in Human Gα Subunits and DYT25 Mutations
(A) Mapping of the dystonia-related mutations on Gαolf sequence.
(B) Structural model of Gαolf built on Gαs crystal structure (PDB: 1AZT) (Figure S2).
(C) Snake plot for human Gαolf, obtained from GPCRdb (Pándy-Szekeres et al., 2018), highlighting residues with natural variants and DYT25 mutations.
(D) Distribution of conservation profiles, calculated as percentage sequence identity for natural and DYT25 mutations.
(E–I) Distribution of PolyPhen (E), SIFT (F), CADD (G), MetaLR (H), and REVEL (I) scores. Statistical significance was assessed using Wilcoxon rank-sum test.
(J) Heatmap showing the prediction of each DYT25 amino acid substitution by different methods, with the bar plot on the right showing the number of methods predicting a given substitution to be deleterious.