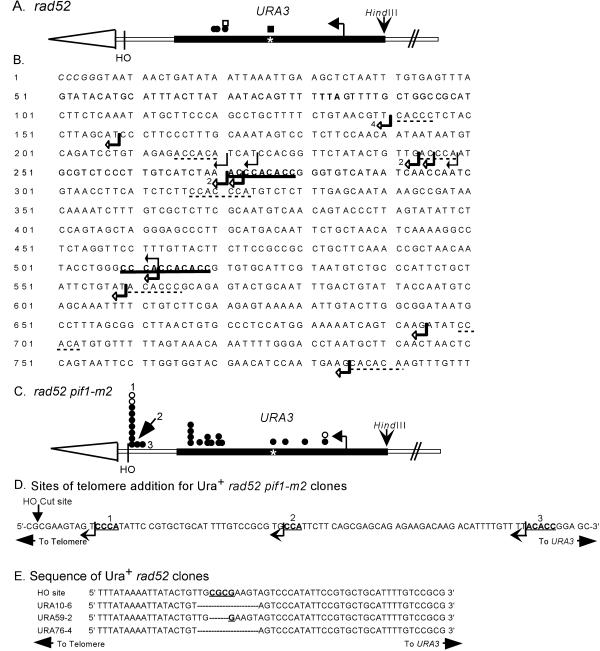
Figure 3.
Positions of de novo telomere addition sites near or within URA3. (A) Positions of independent stabilization events from the rad52 strain are denoted by circles or squares. Circles are clones isolated in the experiment used to determine the frequencies of stabilization reported in Supplementary Table 1A. Squares indicate clones isolated in independent stabilization experiments. Open symbols denote clones whose positions were determined solely by Southern analysis. Closed symbols denote the positions of clones that were mapped precisely by DNA sequencing. The asterisk marks the position of an 11-base pair telomere-like sequence in URA3. The exact sites of telomere addition for these clones are shown in B, which presents the sequence of the strand running 5′ to 3′ from the end toward the center of the chromosome of the URA3 sequence, starting with the SmaI site used to clone the gene. The telomere is distal to the SmaI site. The TAA termination site for Ura3p is in bold. Black arrows indicate the sites of telomere addition in four independent stabilization events isolated in the rad52 strain. Open arrows indicate the sites of telomere addition in 15 independent stabilized clones isolated in the rad52 pif1-m2 strain. In three cases, multiple clones had the same site of telomere addition: in these cases, the number of events at the site is indicated. The 9- and 11-base pair tracts of telomere-like DNA are in bold and underlined. Tracts of telomere-like DNA of 5–7 base pairs are underlined with dotted lines. (C) Positions of independent stabilization events from the rad52 pif1-m2 strain; symbols are the same as in A. The exact sites of telomere addition for these clones are marked by open arrows in B. The positions labeled 1, 2, and 3 are shown in expanded form in D, the sequence of the 100 bps internal to the HO cut site. Telomere addition at these sites yielded Ura+ cells. The arrow indicates the points of transition between the sequence of the test chromosome and the added telomeric DNA. The bold, underlined bases fit the consensus for telomeric DNA. (E) Compares the sequence around the HO cut site in the test chromosome with the sequence for this region in three independent Ura+ stabilized clones isolated in a rad52 strain. The dashes in the sequences indicate missing bases.