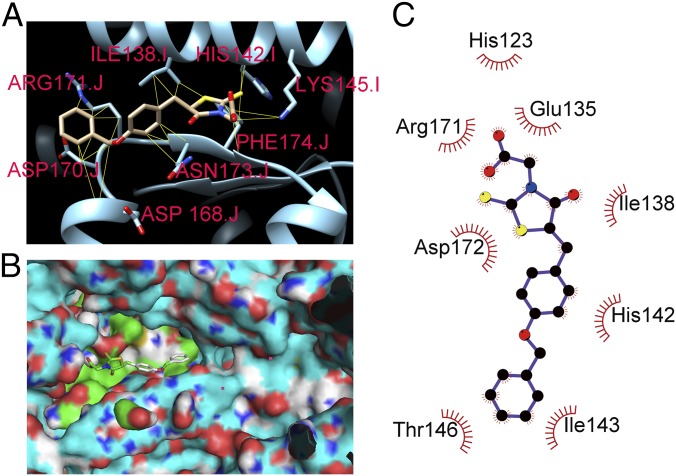
Fig. 3.
Molecular modeling of ClpP-M21 binding. (A) Predicted highest-affinity binding conformation of M21 to ClpP. The ClpP of S. aureus (PDB entry 3v5e) are labeled in cyan. M21 is predicted to bind in close proximity to the catalytically important amino acid residue H142. The side chains of the predicted interacting amino acid residues that participate in the hydrophobic contacts with M21 are shown. (B) Surface representation of ClpP is labeled in cyan. The green color area is the binding pocket, and the cyan area is the active site. (C) 2D schematic diagram of ClpP-M21 interactions is shown. Hydrophobic contacts are represented by an arc with spokes radiating toward the M21 atoms that they contact. The contacted atoms in M21 are shown with spokes radiating back.