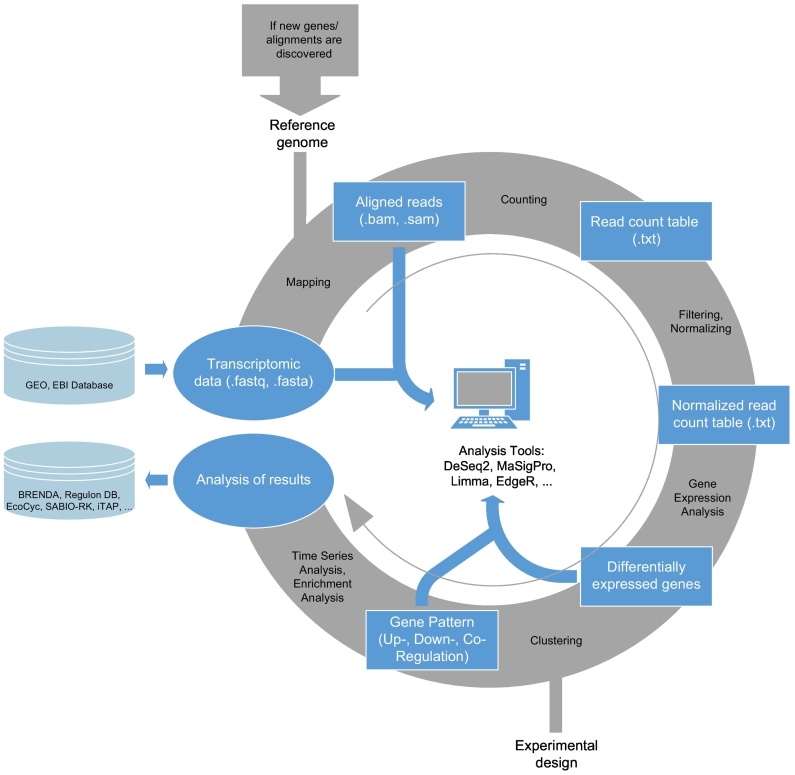
Fig. 2.
Workflow illustrating the general procedure when analyzing gene expression data. RNA Seq analysis creates FASTA and FASTQ as raw data formats compiled in gene expression repositories (GEO, EBI), followed by SAM or BAM files for aligned reads. The analysis steps are, in general: (1) Mapping the reference genome onto the transcriptomic data, (2) Counting reads, (3) Filtering low read counts and normalizing counts, (4) Gene Expression Analysis, (5) Clustering, (6) Time Series Analysis and Enrichment Analysis. DeSeq2, MaSigPro, limma and edgeR are often applied packages within the language R to analyze transcriptomic data, in case of differential gene expression as well as gene pattern analysis. The resulting dynamic expressions and parameters are stored in databases like BRENDA [44], Regulon DB [45], EcoCyc [46], SABIO-RK [47] and iTAP [48].