Fig. 4.
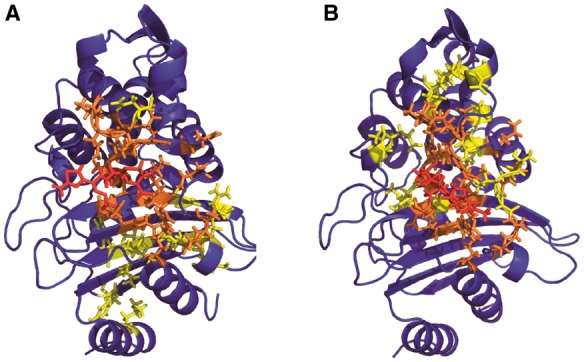
Residues linked to drug motion identified by excess positional mutual information. Residues corresponding to the top 5% of protein atoms scored by excess mutual information to the beta-lactam ring are rendered as sticks on the CTX-M9:cefotaxime structure in panel (A) and the CTX-M9:meropenem structure in panel (B). These residues constitute our predictions for sites where mutation will affect catalytic activity and drug resistance. The drug is shown in red online, residues identified in both enzyme:drug complexes in orange and residues identified in only one enzyme:drug complex in yellow (Color version of this figure is available at Bioinformatics online.)
