Fig.2.
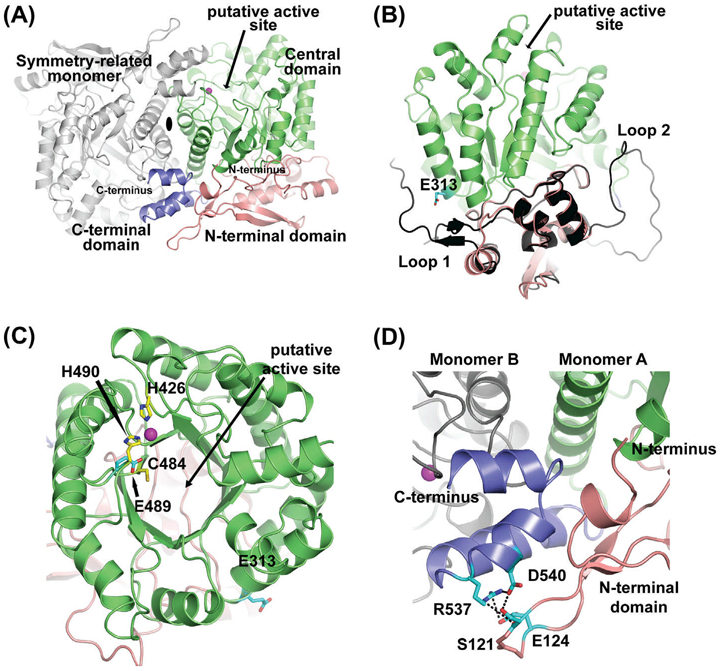
Crystal structure of Arabidopsis THIC. (A) The THIC dimer is shown using a cartoon representation. The dimer is formed by two crystallographically-related molecules around a 2-fold axis (shown as a black ellipse). The N-terminal, central, and C-terminal domains are labeled and colored in salmon, green, and dark blue, respectively. The active site is indicated with an arrow and the position of the metal ion as magenta-colored spheres. The gray-colored molecule is the symmetry-related Arabidopsis THIC monomer. (B) Side view of the Arabidopsis THIC monomer superimposed onto the C. crescentus N-terminal domain structure (colored in black). The two loops present in C. crescentus are absent in the A. thaliana atomic model (labeled as loop 1 and loop 2). Residue E313 is shown as sticks colored according to atom types (carbon cyan, and oxygen red). (C) Top view of the green-colored central domain displaying a typical (β/α)8 TIM barrel fold. The two histidines that partially coordinate the metal ion (H426, H490), one cysteine (C484) involved in the catalytic cycle, the glutamate (E489) implicated in the SAM molecule binding as well as E313 are shown as sticks colored according to atom types (carbon yellow or cyan, oxygen red, and nitrogen blue). (D) Close-up view of the blue-colored C-terminal domain, sandwiched between the N-terminal domain of monomer A colored in salmon and the central domain loops of monomer B in gray. Residues S121, E124, R537, and D540 are shown as sticks and colored to atom types (carbon in cyan, oxygen and nitrogen as before).
