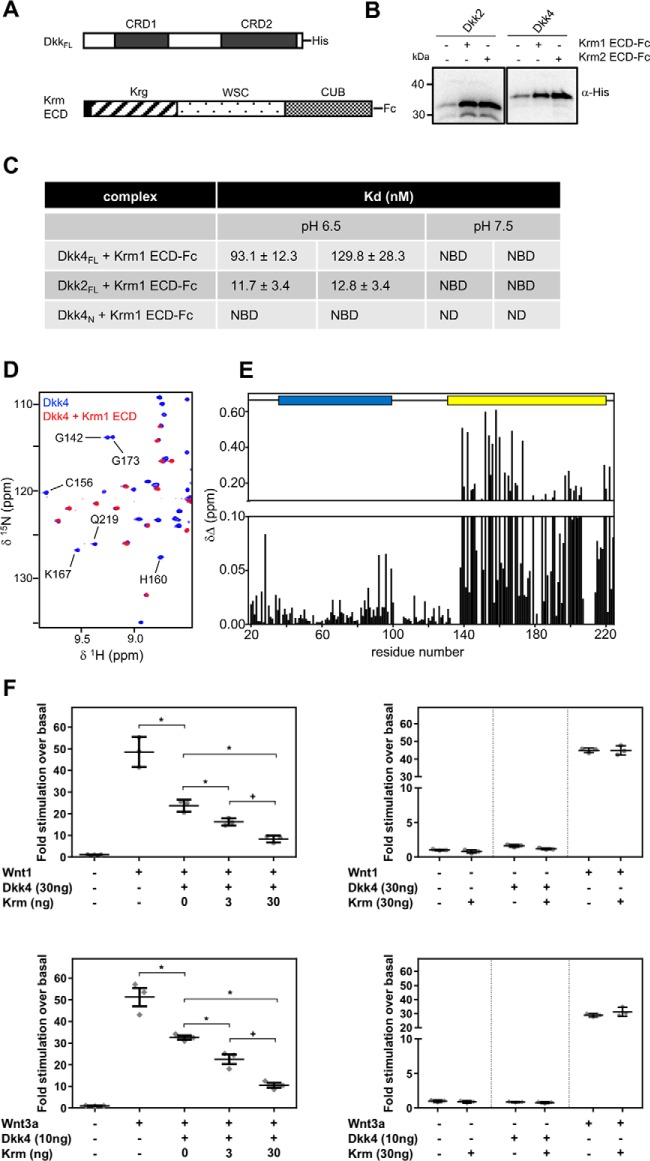
Figure 6.
Dkk2 and Dkk4 bind with high affinity to Krm ECD. A, schematic representations of human His-tagged DkkFL and Krm ECD-Fc constructs. B, Western blot analysis of pulldown assays illustrating the binding of His-tagged DkkFL proteins to Krm1 ECD-Fc and Krm2 ECD-Fc captured on protein A beads. C, a summary of biolayer interferometry KD measurements from two individual experiments and representative of a minimum of three or more independent experiments. Error bars, S.E. calculated for individual fitted curves using Prism version 6.0. NBD, no binding detected; ND, no data obtained. D, overlay of a selected region from the 15N/1H TROSY spectra of uniformly His-tagged 15N/13C/2H-labeled Dkk4FL acquired in the absence (blue) and presence (red) of a 10% molar excess of unlabeled Krm1 ECD. A selection of the well-resolved signals from Dkk4 CRD2 that are lost on binding to the Krm1 ECD are labeled by residue type and number. E, minimum chemical shift perturbation observed for backbone amide groups of His-tagged Dkk4FL induced by the addition of Krm1 ECD. The positions of CRD1 and CRD2 are shown above the histogram as blue and yellow boxes, respectively. F, scatter plots of Wnt reporter assays illustrating synergistic inhibition of Wnt-dependent signaling by Dkk4 and Krm1. HEK293 Tcf-Luc cells were transiently co-transfected with plasmids to express Wnt1 (top left) or Wnt3a (bottom left), together with an amount of plasmid encoding Dkk4 determined to give partial inhibition of Wnt-dependent signaling and increasing amounts of a Krm1 expression plasmid. Appropriate control data are also shown for both the Wnt1 and Wnt3a experiments (top and bottom right, respectively). The results are presented as scatter plots showing individual data points, with bars indicating mean ± S.D. (error bars). Analysis of variance pairwise comparison tests confirm significant differences (p < 0.05) between Dkk4 alone and Dkk4 with both levels of Krm1 (*) and in the responses with different Krm doses (+).