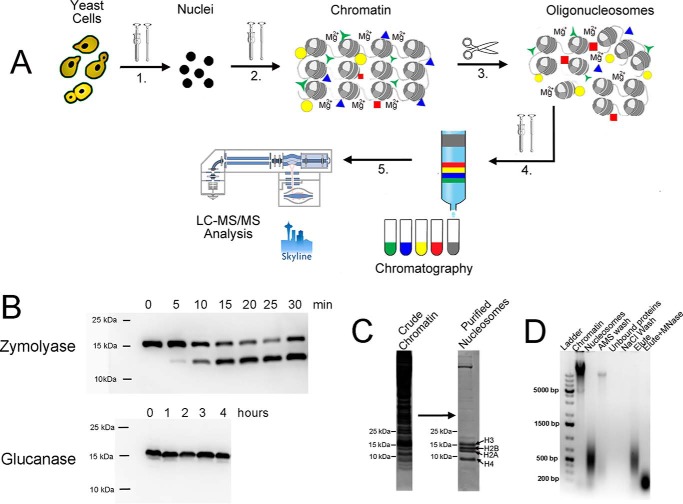
Figure 1.
Optimized chromatin purification protocol using yeast as an example. A, outline of the purification protocol. 1, yeast cell wall was digested using recombinant glucanase, and spheroplasts were disrupted using a Dounce homogenizer. 2, crude nuclei were treated with 400 mm AMS in the presence of 5 mm magnesium chloride. The suspension was homogenized using the Dounce homogenizer, and the bulk chromatin was isolated by centrifugation. 3, oligonucleosomes were generated from bulk chromatin using MNase, sonication, or an EmulsiFlex C5 homogenizer in the presence of 5 mm magnesium chloride (Fig. 2). 4, oligonucleosomes were extracted from the pellet using NaCl and purified using column chromatography (Fig. 3). 5, purified nucleosomes were isolated and analyzed using LC-MS/MS (Fig. 4). B, immunoblotting reveals H3 degradation (clipping) in the presence of zymolyase, but not glucanase, during cell wall digestion. C, Coomassie-stained polyacrylamide gel of yeast chromatin before and after purification. D, ethidium bromide–stained DNA-agarose gel of yeast chromatin purification procedure.