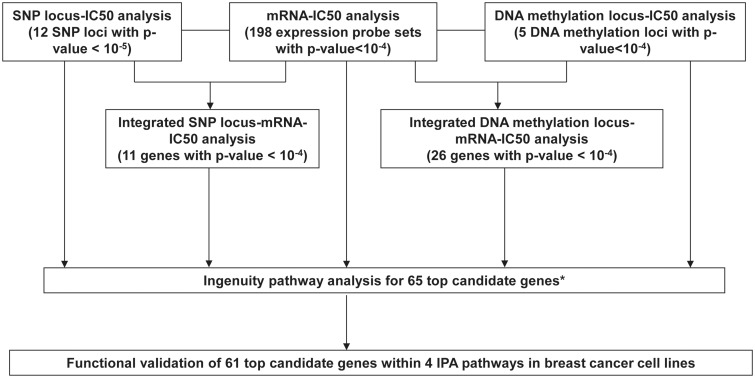
Figure 2.
Schematic diagram of the strategy used to select genes for functional validation. Genome-wide association studies were performed using metformin IC50 with 1.3 million SNPs, 54K expression probe sets or 485K DNA methylation probes. Integrated SNP/Methylation-Expression-IC50 analyses were performed using SNP loci that contained at least 1 SNP with P-value < 10−5 and 1 SNP with P-value < 10−3, or DNA methylation loci that located in the expressed gene region and contained at least two probes associated with metformin IC50 with P-value <10−4, 54K expression probe sets and metformin IC50 to identify SNP loci/methylation loci associated with metformin IC50 through their influence on gene expression. Ingenuity pathway analysis (IPA) was performed for the top 65 candidate genes. Finally 61 genes within 4 IPA networks were selected for functional validation studies using siRNA screening approach in breast cancer cell lines. * One gene, CDC42BPA, was also included in the top 65 candidates, which contained one methylation probe associated with its own gene expression and metformin IC50 with P-value < 10−4, and which gene expression was associated with metformin IC50 with P-value < 10−2.