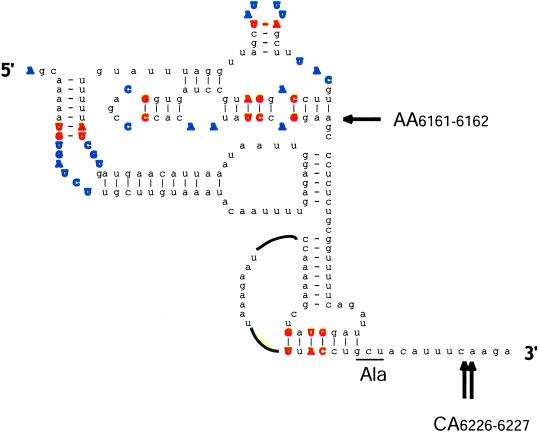
Figure 1.
Predicted structure of the CrPV IGR-IRES. Base pairs conserved among the IGR IRESs of cricket paralysis-like viruses and unpaired residues are shown in bold. The underlined GCU triplet is decoded ribosomal A site into the N-terminal alanine of the protein encoded by the second ORF of the viral mRNA. Arrows mark nucleotides where the leading edge of the ribosome was mapped by the technique of “ribosomal toeprinting” in 40S/IGR IRES complexes. Only the toeprints at residues 6226 and 6227 correlated with the ability of the IRES to function in internal initiation (21). The drawing was provided courtesy of C. Hellen (State University of New York, Brooklyn).