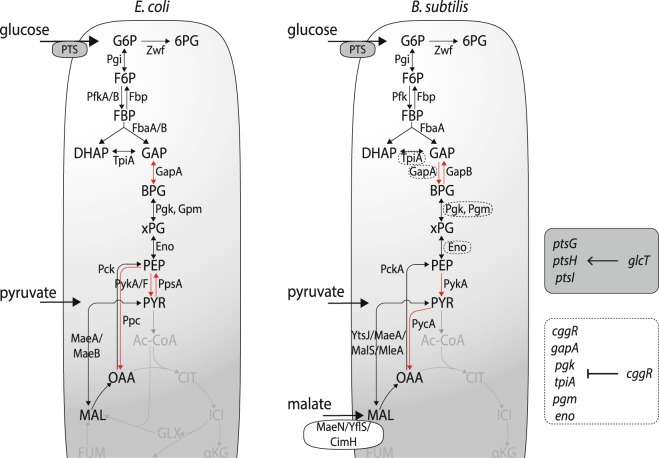
Figure 1.
Glycolysis in E. coli and B. subtilis. Differences between the two organisms are highlighted in red. Metabolites: G6P = glucose 6-phosphate, F6P = fructose 6-phosphate, FBP = fructose 1,6-biphosphate, DHAP = dihydroxyacetone phosphate, GAP = glyceraldehyde 3-phosphate, BPG = 1,3-biphosphoglycerate, xPG = 2/3-phosphoglycerate, PEP = phosphoenol pyruvate, PYR = pyruvate, OAA = oxaloacetate, MAL = malic acid. Enzymes: Zwf = G6P dehydrogenase, Pgi = G6P isomerase, Pfk = 6-phosphofructokinase, Fbp = fructose-1,6-biphosphatase, Fba = FBP aldolase, Tpi = triose phosphate isomerase, Gap = GAP dehydrogenase, Pgk = phosphoglycerate kinase, Pgm = phosphoglycerate mutase, Eno = enolase, Pyk = pyruvate kinase, Pck = PEP carboxykinase, YtsJ = NADP-dependent malic enzyme, Mae = NAD-dependent malic enzyme, MalS = NAD-dependent malic enzyme, MleA = NAD-dependent malic enzyme, Pps = PEP synthetase, Pyc = PYR carboxylase, Ppc = PEP carboxylase. Transport: PTS = phosphotransferase system, ptsG = phosphotransferase system (PTS) glucose-specific enzyme IICBA component, ptsH = polypeptide: histidine-containing phosphocarrier protein of the phosphotransferase system (PTS) (HPr protein), ptsI = phosphotransferase system (PTS) enzyme I. Regulators: CggR = negative regulator of eno-operon, glcT = transcriptional antiterminator (BgIG family)