Abstract
Circular RNAs (circRNAs) are a series of special closed circular RNA molecules with stability and conservatism. In recent years, advances in high-throughput RNA sequencing technology have led to explosive discovery of circRNAs in different types of species and cells. Moreover, circRNAs can accomplish a remarkable multitude of biological functions, such as regulating transcription or splicing, serving as miRNA sponges, interacting with RNA-binding proteins, and translating proteins. Meanwhile, circRNAs involve in the biogenesis and development of many diseases, including cardiovascular disorders, nervous system disorders, cancers, etc. Herein, we discuss the latest research progress of circRNA, as well as their diagnostic and prognostic significance in digestive system cancers. In addition, this paper highlights that circRNAs might serve as potential therapeutic targets for novel drugs by taking digestive system cancer as an illustrative example.
Keywords: Circular RNA, digestive system cancer, diagnosis, prognosis, therapeutic targets
Introduction
Circular RNA (circRNA) is a special single-stranded closed circular RNA molecule in nature. It was first found in viroid as early as in 1976 [1], and was later isolated from several eukaryotes in 1979 [2]. Its biological significance kept unknown until scientists gradually unveiled its mystery with the help of high-throughput sequencing technology and bioinformatics in the 21st century. And the researches on circRNAs have seen explosive growth since 2012. Using innovative methods (for example, RNA-seq [3], RPAD [4], and circBase [5]), scientists have identified thousands of circRNAs in human beings [6], plants [7], and some animals [6,8,9]. Unlike linear RNAs, circRNAs, as a result of their special structure, are more stable, conserved, highly abundant, and dynamically expressed in a spatio-temporal manner inside particular tissues [6,8-12]. Because of these biological properties, circRNAs may play significant roles as special biomarkers in occurrence and development of diseases. In recent years, circRNAs have gradually been demonstrated to correlate with various human diseases, including cardiovascular disorders [13-15], nervous system disorders [16,17], intervertebral disc degeneration [18], silicosis [19], diabetic retinopathy [20], and cancers [21-26]. They also can serve as novel therapeutic targets for above diseases [14,16,18,27]. This review comprehensively and concisely summarizes the biogenesis and function of circRNAs, and describes the application of main circRNA databases. Furthermore, we mainly discuss the emerging roles of circRNAs in digestive system cancers. Finally, the paper especially highlights that circRNAs might serve as potential therapeutic targets for novel drugs by taking digestive system cancers as an illustrative example.
The biogenesis of circRNAs
Unlike linear RNAs generated from canonical splicing, circRNAs usually arise from back-spliced events of precursor mRNAs (pre-mRNAs) and circularization of exons, introns, or both exons and introns [28]. Besides, canonical pre-mRNA splicing can compete against exon circularization in a tissue-specific and conserved manner from flies to humans [29-31]. Typically, a circular RNA splice junction is formed by a donor site at a downstream 3’ end joining with an acceptor splice site at the 5’ end of an exon (head-to-tail) [6,32]. In human cells, ecircRNAs (exonic circRNAs) contain one or more exons. They account for the main part of circRNAs, and play an important role in many complicated yet critical biological functions [6,33,34]. One mechanism of ecircRNAs biogenesis is exon circularization mediated by flanking intronic complementary sequences. Noticeably, the formation of reversely repeated Alu pairs and the competition between different Alu pairs can lead to alternative circularization, producing multiple circular RNA transcripts from a single gene (Figure 1A) [35]. In another mechanism, ecircRNAs biogenesis can be regulated by RNA-binding proteins (RBPs) (Figure 1B) [29,30]. Taking circMbl as an example, muscleblind (MBL), as a regulatory factor, can promote circMbl biogenesis. It depends on the presence of functional MBL binding sites in the flanking intronic sequences [29]. Quaking (QKI) is another RBP that strikingly facilitates circRNA formation via pre-mRNA binding sites. During human epithelial-mesenchymal transition (EMT), over one third of circRNAs abundance is dynamically regulated by QKI. It is dependent on intronic QKI binding motifs [30]. In addition, scientists have also identified three other special subclasses of circRNAs with different biogenesis mode: circular intronic RNAs (ciRNAs) [36], exon-intron circRNAs (EIciRNAs) [37], and tRNA intronic circRNAs (tricRNAs) [31]. Unlike ecircRNAs, EIciRNAs retain the introns instead of removing them during splicing process. Moreover, some pre-mRNAs contain flanking Alu complementary pairs, while others contain flanking complementary sequence pairs other than Alu. Both of them facilitate the production of EIciRNAs (Figure 1A) [37]. Interestingly, ciRNAs derive from introns and accumulate due to a failure in debranching. Their formation depends on a consensus motif containing a 7 nt GU-rich element near the 5’ splice site and an 11 nt C-rich element close to the branchpoint site (Figure 1C) [36]. TricRNAs are a special type of intronic circRNAs generating during pre-tRNA splicing [31]. Their expression is regulated in an age-dependent and tissue-specific manner. Biogenesis of tricRNAs requires anciently conserved tRNA sequence motifs and processing enzymes.
Figure 1.
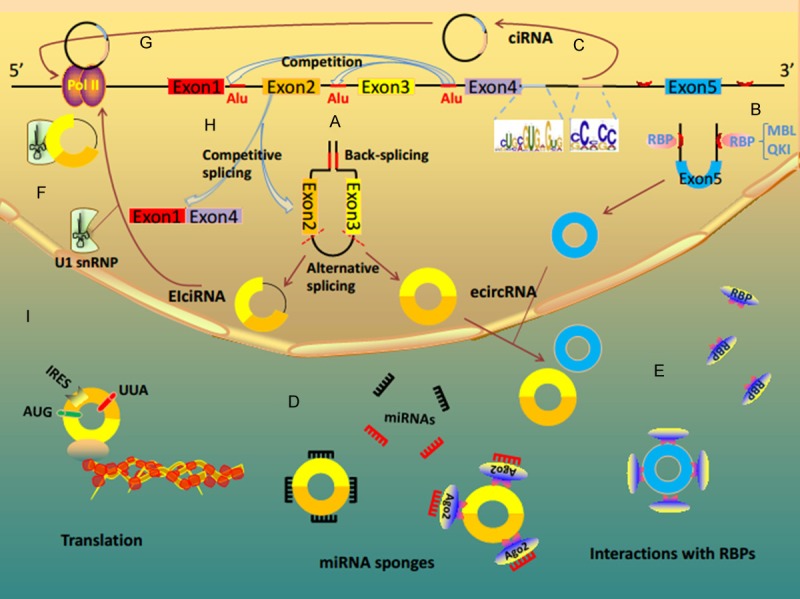
CircRNA biogenesis and function. A. The formation of reversely repeated Alu pairs and the competition between different Alu pairs leading to alternative circularization. During the procession of circularization, pre-mRNAs form ecircRNA or EIciRNA by removing or retaining intron respectively. B. EcircRNA biogenesis can be regulated by RNA-binding proteins, such as MBL and QKI. C. CiRNAs are derived from introns. They depend on a consensus motif containing a 7 nt GU-rich element near the 5’ splice site and an 11 nt C-rich element close to the branchpoint site. D. CircRNAs serve as miRNA sponges to inhibit miRNA activity. E. Some circRNAs function as protein sponges and interact with RNA binding proteins (RBPs). F. Some EIciRNAs regulate Pol II transcription of their parental genes via specific RNA-RNA interaction between U1 snRNA and EIciRNAs. G. Some ciRNAs function as positive regulators of Pol II transcription and play a vital part in the efficient transcription of their parent coding genes. H. CircRNAs can modulate linear splicing by competing for splice sites of pre-mRNA. The formation of circRNA affects the alternative splicing of pre-mRNA, generating circRNA and corresponding linear RNA. I. Some circRNAs have protein-coding capacity and can translate proteins.
Function of circRNAs
As miRNA sponges: yes or no?
The hallmark function of circRNAs is their ability to serve as miRNA sponges (Figure 1D). The first observation of a circRNA acting as a miRNA sponge is ciRS-7 (also termed as CDR1). It has more than 70 conserved binding sites of miRNA-7. Thus, ciRS-7 likely plays profound roles in diseases such as cancer by fine-tuning the miR-7/miR-671/ciRS-7 axis [6,38,39]. Analogously, experiments with circular SRY RNA uncovered its function as a miR-138 sponge [39]. Since then, there have been more and more studies on the function of circRNAs as miRNA sponges. For example, circ-Foxo3, totally with 25 miRNA-binding sites, functions as a sponge for eight miRNAs. The ectopic expression of Foxo3 circRNA could suppress tumor growth, cancer cell proliferation and survival [40]. Similarly, circHIPK3 serves as a modulator of cell growth by sponging multiple miRNAs (9 miRNAs with 18 binding sites) in human cells [11]. These results indicate that one circRNA might be associated with a variety of miRNAs. But unlike ciRS-7, circHIPK3 only have 1-5 binding sites for one miRNA. Consistent with previous findings, circRNA HRCR was also proved to serve as a miR-223 sponge to regulate cardiac hypertrophy and heart failure [14]. Maybe it is a general phenomenon for circRNAs served as miRNA sponges.
However, as a special novel type of endogenous noncoding RNA, controversy on circRNAs is inevitable. For instance, Jeck et al. [41] showed that very few circRNAs in mammalian cells contained more than ten binding sites for an individual miRNA. Many exonic circRNAs just contained a smaller number of putative miRNA binding sites. Soon afterwards, Guo et al. [3] and Militello et al. [42] argued against the generalized properties that circRNAs act as miRNA sponges. Instead, they all held a view that most circRNAs do not function as miRNA sponges. Since most circRNAs are of low abundance and in small length, many circRNAs may not serve as miRNA sponges [11]. Therefore, it still remains to be illustrated whether miRNA sponges are widespread regulators of miRNA activity in eukaryotes, and how networks of circRNAs, miRNAs, and ceRNAs regulate cellular homeostasis.
Interaction with RBPs
In addition to the function of miRNA sponges, some circRNAs have a high density of binding sites for a single or multiple RBPs. They may function as protein sponges interacting with RBPs (Figure 1E) [43]. A typical example is ciR-7/CDR1as, widely associating with Argonaute (AGO) proteins in a miR-7-dependent manner [39]. Likewise, due to the presence of functional MBL binding sites in circMbl, MBL can directly bind to circMbl and promote circMbl production. Downregulation of MBL in both cultured cells and fly neural tissue leads to a remarkable decrease in circMbl production [29]. Another study revealed a competitive phenomenon between a circRNA and its homologous RBP mRNA. The extensive binding of circPABPN1 to HuR prevents HuR binding to PABPN1 mRNA and lowers PABPN1 translation [44]. Circ-Foxo3, encoded by the Foxo3 gene [45], appears to have tumor suppressive activity like its parental gene. Circ-Foxo3 is able to bind with 5 types of proteins including cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21) with a high affinity. Ectopic expression of circ-Foxo3 can suppress cell proliferation and block cell cycle progression, because it can bind to the cell cycle proteins CDK2 and p21 to form a circ-Foxo3-p21-CDK2 ternary complex [46].
Regulating transcription and splicing
EIciRNAs, predominantly localized in the nucleus, may have the function of gene transcription regulation [28,36,37]. For example, circEIF3J and circPAIP2 might regulate the Pol II transcription of their parental genes via specific RNA-RNA interaction between U1 snRNA and EIciRNAs (Figure 1F) [37]. CiRNAs, another subclass of circRNAs, can also regulate gene expression. They are abundant in the nucleus but have few miRNA target sites. Some ciRNAs, such as ci-ankrd52 and ci-sirt7, function as positive regulators of Pol II transcription and play an important role in the efficient transcription of their parent coding genes (Figure 1G) [36].
CircRNAs also modulate linear splicing by competing with splice sites of pre-mRNA. Thus, the formation of circRNA affects the alternative splicing of such pre-mRNA, potentially generating circRNA and corresponding linear RNA (Figure 1H) [29,34]. As previously described, MBL can bind directly to circMbl and promote circMbl production. Thus, it negatively affects canonical splicing and decreases the production of the parental mRNA [29]. Additionally, abundant ecircRNAs derive from exon 2 of HIPK2/3 undergoing “alternative” splicing to circularize rather than canonical splicing to produce linear transcript. Circularization of HIPK2/3 negatively regulates the usual protein-encoding transcript production [12].
Translating proteins
Translating proteins is another amazing discovery about the biological function of circRNAs (Figure 1I), which is contrary to the conventional concept of circRNAs [32,47]. Previously, protein-coding process was thought to merely transfer from DNA to a linear mRNA then to a protein. CircRNA was regarded as a distinct class of endogenous noncoding RNA, because it is lack of a 5’ 7-methylguanosine cap structure as well as a poly (A) tail. Both of them are required for efficient translation of linear mRNAs [48]. But as early as 1995, eukaryotic ribosomes was reported to initiate translation on circRNAs in vitro, as RNAs contain internal ribosome entry site (IRES) elements [49]. Besides, covalently closed circular (CCC) RNA is the smallest (220 nt) RNA among all known viroids and virusoids. In 2014, the CCC RNA was found to contain an IRES element and was directly translated by eukaryotic ribosomes through two (or three) completely overlapping open reading frames (ORFs) [50]. Since then, more and more convincing studies have emerged that circRNAs may be used as templates for protein synthesis [51-57].
So far, at least four studies have offered sufficient proofs for the existence of translatable circRNA molecules: circMbl [56], circ-ZNF609 [57], circ-FBXW7 [53], and circ-SHPRH [54]. Circ-ZNF609 contains an ORF spanning from the start codon and terminating at an in-frame STOP codon. It can be translated into a protein in a splicing-dependent and cap-independent manner [57]. Analogously, circMbl can also be translated in a cap-independent way in drosophila heads. Furthermore, starvation and FOXO likely regulate the translation of a circMbl isoform [56]. Additionally, circ-SHPRH [54] and circ-FBXW7 [53], as well as proteins coded by them, are found to be abundantly expressed in normal human brains while down-regulated in glioma. Both of the circRNAs have an ORF driven by the IRES to translate a functional protein. The translation from circRNA may be prevalent in other cancers, as a question to be addressed in the future.
Different from these individual examples, N6-methyladenosine (m6A), the most abundant base modification of RNA, was reported to promote efficient initiation of protein translation from circRNAs [55]. The phenomenon is widespread in human cells. Hundreds of endogenous circRNAs have translation potential. Meanwhile, circRNA translation initiated by m6A is heightened under heat shock condition. It is indicated that circRNA-encoded proteins may play important roles in cellular responses to environmental stress.
All of the above researches suggested that endogenous circRNAs may generate proteins. It expands the coding landscape of human transcriptome, and provides a new direction for circRNA research in the future.
CircRNA databases
With the application of next-generation sequencing and bioinformatics, numerous circRNAs are being discovered. They are integrated into the circRNA database for more thorough researches (Table 1). These databases contain a mass of circRNA genomes and mature RNA sequences of different species, as well as circRNAs relating to various diseases [5,58-62]. Hence, they provide a transparent and comprehensive view of the spatio-temporal presence and function of circRNAs. Some databases are capable of detecting circRNAs with protein-coding potential [63,64], while others are able to predict interaction networks between noncoding RNAs, circRNAs, and RBPs [58-60,65,66].
Table 1.
Databases of circRNAs
| Database | URL | Description | Ref. |
|---|---|---|---|
| CircBase | http://www.circbase.org/ | Provides merged and unified datasets of circRNAs, with evidence supporting their expression | [5] |
| CircNet | http://circnet.mbc.nctu.edu.tw/ | Provides information on known and novel circRNAs, circRNA-miRNA-gene regulatory networks | [66] |
| CircInteractome | https://circinteractome.nia.nih.gov/ | Provides bioinformatic analyses of binding sites on circRNAs, identifies potential circRNAs acting as RBP sponges and potential internal ribosomal entry sites, designs primers or siRNAs for specific circRNAs | [65] |
| CircRNADb | http://reprod.njmu.edu.cn/circrnadb/ | Annotates protein-coding potential of each circRNA | [64] |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | Predicts interactions among the miRNAs, protein coding, long non-coding and circular RNA genes, checks the enrichment of genes associated with particular biological processes by analyzing protein coding genes in the miRNA-circRNA interactome of individual diseases, and identifies disease associated SNPs on circRNA loci and Argonaute (Ago) interaction sites on circular RNAs | [60] |
| starBase v2.0 | http://starbase.sysu.edu.cn/ | Predicts interaction networks between lncRNAs, miRNAs, circRNAs, mRNAs, and RBPs from large-scale CLIP-seq data | [59] |
| deepBase v2.0 | http://deepbase.sysu.edu.cn/ | Provides comprehensive expression and evolution profiles of lncRNAs, circRNAs and small RNAs, identifies and annotates lncRNAs and circRNAs | [58] |
| CIRCpedia | http://www.picb.ac.cn/rnomics/circpedia/ | Identifies and annotates back-splicing and alternative splicing of circRNAs in different cells | [111] |
| TSCD | http://gb.whu.edu.cn/TSCD/ | Provides a new integrated view to explore the tissue-specific circRNAs in human and mouse tissues | [61] |
| CSCD | http://gb.whu.edu.cn/CSCD | Provides an integrated circRNA database to promote functional studies of cancer-specific circRNAs | [62] |
| CircPro | http://bis.zju.edu.cn/CircPro | An integrated tool capable of detecting circRNAs with protein-coding potential from high-throughput sequencing data | [63] |
Recent research progress of circRNAs in digestive system cancer
CircRNAs widely exist in normal human tissues and cells. They possess a variety of biological functions to regulate gene expression in a spatio-temporal and tissue-specific manner. The translation of circRNAs can be regulated under stress conditions [55,56]. Thus, circRNAs might serve as potentially novel and promising biomarkers for the diagnosis, therapy, and prognosis of human diseases, especially cancer. This review mainly focuses on the recent research progress in digestive system cancer. The detailed information is summarized in Table 2.
Table 2.
CircRNAs in digestive system cancer: signaling pathway and function
| circRNA | Cancer type | Expression in tumors | Pathway/regulatory network | Function/clinical association | Ref. |
|---|---|---|---|---|---|
| cir-ITCH | ESCC | Low | cir-ITCH-miR-7/miR-17/miR-214-ITCH-Wnt signaling | Reduces cell viability and arrests proliferation in ESCC cells | [67] |
| Colorectal cancer | Low | cir-ITCH-Wnt signaling | Plays an anti-tumor role | [78] | |
| HCC | Low | Not mentioned | Has an inhibitory effect on HCC, and associates with survival of HCC | [112] | |
| hsa_circ_006793 | ESCC | High | Not mentioned | Correlates with tumor differentiation and TNM stage; promotes proliferation and migration of ESCC cell | [68] |
| circPVT1 | Gastric cancer | High | circPVT1-miR-125-E2F2 | An independent prognostic factors for overall survival and disease free survival in patients with gastric cancer; affects proliferation of gastric cancer cells | [24] |
| ciRS-7 (CDR1as) | Gastric cancer | High | ciRS-7-miR-7-PTEN/PI3K/AKT pathway | An independent risk factor of overall survival; Correlates with a more aggressive oncogenic phenotype | [73] |
| Colorectal cancer | High | ciRS-7-miR-7-EGFR/RAF1/MAPK pathway | An independent risk factor for overall survival with tumor stimulative effect | [79] | |
| HCC | high | ciRS-7-miR-7-EGFR or PI3K/AKT/mTOR pathway | An independent risk factor of hepatic microvascular invasion; correlates with the proliferation of hepatocellular carcinoma cells and the deterioration of HCC | [83,84] | |
| hsa_circ_0001895 | Gastric cancer | Low | Not mentioned | Associates with cell differentiation, Borrmann type, and tissue CEA expression; has a better sensitivity and specificity than the commonly used biomarkers of gastric cancer | [74] |
| circLARP4 | Gastric cancer | Low | circLARP4-miR-424-LATS1-YAP pathway | An independent prognostic marker for overall survival of gastric cancer patients as well as the patients with chemotherapy; inhibits DNA synthesis, cell proliferation and invasion; correlates with tumor size and lymphatic metastasis | [75] |
| hsa_circ_0000745 | Gastric cancer | Low | hsa_circ_0000745-miRNAs | Associates with tumor differentiation; has higher sensitivity and specificity than serum CEA levels | [76] |
| hsa_circ_000984 | Colorectal cancer | High | hsa_circ_000984-miR-106b-CDK6 | Promotes colorectal cancer cell proliferation, migration, invasion and tumor formation; correlates with advanced colorectal cancer | [81] |
| hsa_circ_001569 | Colorectal cancer | High | hsa_circ_001569-miR-145-E2F5/BAG4/FMNL2 | Promotes colorectal cancer cell proliferation and invasion | [82] |
| circCCDC66 | Colorectal cancer | High | circCCDC66-miRNAs-oncogenes | Promotes colorectal cancer cell proliferation, migration, invasion, and anchorage-independent growth; associates with poor prognosis | [22] |
| circHIPK3 | Colorectal cancer | High | c-Myb-circHIPK3-miR-7-FAK, IGF1R, EGFR, YY1 | Positively correlated with metastasis and advanced clinical stage; an independent prognostic factor of poor overall survival in colorectal cancer | [80] |
| circMTO1 | HCC | Low | circMTO1-miR-9-p21 | Suppresses hepatocellular carcinoma cell proliferation and invasion; associates with survival of HCC | [26] |
| hsa_circ_0004018 | HCC | Low | hsa_circ_0004018-miR-30e-5p/miR-626-MYC | Correlates with serum AFP level, tumor diameters, differentiation, BCLC stage and TNM stage; HCC-stage-specific expression features in diverse chronic liver diseases | [85] |
| circZKSCAN1 | HCC | Low | circZKSCAN1-miRNA-Mrna | Associates with tumor numbers, cirrhosis, vascular invasion, or microscopic vascular invasion; inhibits growth, migration, and invasion of HCC | [86] |
| cSMARCA5 | HCC | Low | cSMARCA5-miR-17-3p/miR-181b-5p-TIMP3 | An independent risk factor for overall survival and relapse-free survival after hepatectomy; inhibits the proliferation and migration of hepatocellular carcinoma cells | [87] |
| circRNA_100338 | HCC | High | circRNA_100338-miR-141-3p | Correlates with a low cumulative survival rate and metastatic progression in HCC patients with hepatitis B | [88] |
| circARSP91 | HCC | Low | AR-ADAR1-CircARSP91 | Suppresses tumor growth | [89] |
Esophageal cancer
By analyzing a total of 684 esophageal squamous cell carcinoma (ESCC) and paired adjacent non-tumor tissue samples, researchers found that cir-ITCH may have an inhibitory effect on ESCC by regulating the Wnt pathway [67]. On the contrary, another study indicated that hsa_circ_0067934 was able to promote the proliferation and migration of ESCC cell lines [68]. Its expression was associated with tumor differentiation and tumor-node-metastasis (TNM) stage. Besides, the abnormal expression of circRNAs may play a role in radiotherapy resistance of esophageal cancer [69]. Recently, Sun et al. [70] provided the first global circRNA expression profiling of all the types of esophageal tumor cells by a combination of RNA sequencing and bioinformatics analysis. They found that circRNAs were involved in tumor metabolism, cell apoptosis, proliferation, and migration.
Gastric cancer
Gastric cancer is one of the commonest malignancies worldwide [71]. It lacks reliable and efficient early diagnostic biomarkers, therefore the 5-year overall survival (OS) rate remains low in advanced patients [72]. To improve the clinical outcome of patients, it’s essential to identify effective biomarkers for early diagnosis. Several studies revealed that circRNA may act as a prognostic marker in gastric cancer. CircPVT1, for example, is a novel proliferative factor and prognostic marker in gastric cancer. It was upregulated in patients with gastric cancer [24]. But surprisingly, high levels of circPVT1 predicated a significantly better overall survival in gastric cancer patients than those with low levels of circPVT1. It may be explained by the positive association of circPVT1 with the tumor suppressor miR-125. Analogously, ciRS-7 was significantly upregulated in gastric cancer tissues compared with the normal tissues. The upregulation of ciRS-7 was linked to poor survival and was probably an independent risk factor of overall survival [73]. Overexpression of ciRS-7 in MGC-803 and HGC-27 cells led to a more aggressive oncogenic phenotype through resisting miR-7-mediated PTEN/PI3K/Akt pathway with tumor-suppression effect. Recently, three independent studies respectively found that hsa_circ_0001895 [74], circLARP4 [75], and hsa_circ_0000745 [76] were relatively downregulated in gastric cancer tissues compared to adjacent noncancerous tissues. Hsa_circ_0001895 was decreased not only in gastric cancer tissues but also in gastric precancerous lesions [74]. It was significantly correlated with cell differentiation, Borrmann type, and tissue carcinoembryogenic antigen (CEA) expression. CircLARP4 inhibited biological behaviors of gastric cancer cells by sponging miR-424. It represented an independent prognostic factor for overall survival of gastric cancer patients [75]. Hsa_circ_0000745 was also down-regulated in plasma samples from patients with gastric cancer. The expression level in gastric cancer tissues and plasma was correlated with tumor differentiation and TNM stage respectively. Additionally, combined with CEA level, plasma hsa_circ_0000745 level is a more promising diagnostic marker for this malignancy [76]. Interestingly, a four-circRNA-based classifier model, established by Zhang et al., could predict the early recurrence of stage III gastric cancer after radical surgery. Its accuracy can be further improved by combining with traditional TNM staging [77]. In conclusion, circRNAs play crucial roles during gastric cancerogenesis and are potential biomarkers for clinical prognosis prediction. What’s more, tissue-specific expression of circular RNAs combined with the commonly used biomarkers may contribute to improving the sensitivity and specificity in the screening of gastric cancer.
Colorectal cancer
During the development of colorectal cancer, some circRNAs serve as oncogenes, while others function as anti-oncogenes. For instance, cir-ITCH was down-regulated in colorectal cancer compared with the peritumoral tissue and was related with Wnt/β-catenin pathway [78]. However, ciRS-7 was obviously upregulated in colorectal cancer. Its overexpression inhibited miR-7 and subsequently activated EGFR and RAF1 oncogenes. CiRS-7 emerged as an independent risk factor for overall survival [79]. More recently, circHIPK3 was found to be significantly upregulated in colorectal cancer tissues and cell lines [80]. It positively correlated with metastasis and advanced clinical stage. Moreover, high-level expression of circHIPK3 was an independent prognostic factor of poor overall survival in colorectal cancer. There are also several other circRNAs serving as oncogenes in colorectal cancer, such as hsa_circ_000984 [81], hsa_circ_001569 [82], and circCCDC66 [22]. On one hand, all of them promoted colorectal cancer cells proliferation, invasion [82], and even metastasis [22,81]. On the other hand, they exerted functions via circRNA/miRNA/targets axis [81,82] or directly regulating a subset of oncogenes [22].
Hepatocellular carcinoma (HCC)
HCC is the commonest malignancy of liver leading to death due to high frequency of metastasis and recurrence [71]. It is urgent to identify potential biomarkers and to find new targets for designing more effective therapies. Recent years, numerous miRNAs and long noncoding RNAs (lncRNAs) have been found to closely relate with the occurrence of HCC. Though the regulation and function of circRNAs in HCC remains largely unknown, there is mounting evidence that circRNAs are involved in HCC development.
CiRS-7 (also termed as CDR1as) is the best-known circRNA so far functioning as a sponge for miR-7 [6]. It has been mentioned in gastric cancer and colorectal cancer above. Researchers found that the overexpression of ciRS-7 in HCC was significantly correlated with hepatic microvascular invasion (MVI), alpha fetal protein (AFP) level, and younger age, thus it might be related with the deterioration of HCC [83]. Subsequently, ciRS-7 was revealed to exert its effects on cell proliferation by regulating epidermal growth factor receptor (EGFR) expression via targeting miR-7 in HCC cells [84]. Additionally, several circRNAs with differential expression have been identified in liver cancer. Taking circMTO1 as an example, it was significantly down-regulated in HCC [26]. CircMTO1 held back HCC progression through acting as the sponge of oncogenic miR-9 to promote p21 expression. Low-level expression of circMTO1 in HCC tissues may be a prognosis predictor for shortened survival of patients. Similarly, hsa_circ_0004018 also decreased obviously in HCC tissues [85]. It was correlated with serum AFP level, tumor diameters, differentiation, Barcelona Clinic Liver Cancer (BCLC) stage, and TNM stage. More importantly, hsa_circ_0004018 showed stage-specific expression features in diverse chronic liver diseases from chronic hepatitis, cirrhosis to HCC. It may play an important part in the carcinogenesis and metastasis of HCC. Interestingly, another study manifested that ZKSCAN1 gene and its related circRNA (circZKSCAN1) inhibited HCC cell growth, migration, and invasion through different downstream signaling pathways [86]. Recently, circRNA cSMARCA5 was found to promote the expression of TIMP3, a well-known tumour suppressor, via sponging miR-17-3p and miR-181b-5p [87]. It could inhibit the proliferation and migration of HCC. Furthermore, the down-regulation of cSMARCA5 in HCC was strongly associated with invasion. It might serve as an independent risk factor for overall survival and recurrence-free survival (RFS) after hepatectomy in HCC patients. In addition, circRNA_100338, highly expressed in HCC, was closely correlated with a low cumulative survival rate and a high metastasis rate in HCC patients with hepatitis B [88]. Finally, androgen receptor (AR), a transcriptional activator of ADAR1 promoter, was found to suppress circRNA expression by upregulating ADAR1. More significantly, ADAR1 expression could effectively predict prognosis of HCC patients and was positively correlated with AR in HCC. At the same time, circARSP91, downregulated by AR in an ADAR1-dependent manner, could inhibit HCC tumor growth [89]. The results provided an explanation about the obvious gender bias of HCC from a new angle. The rapidly growing interest and emerging data are likely to provide new insights for the research of circRNA and help to identify novel candidates for biomarkers or therapeutic intervention.
Therapeutic approaches targeting circRNAs in digestive system cancer
Differential expression of circRNA has been reported in tumors compared with normal tissues. The identification of circRNA contributing to tumor suppression or tumorigenesis provides opportunities to develop novel therapeutics for cancer [11,46,53]. The relationship between various novel circRNAs and carcinogenesis signaling pathways indicates that circRNAs are potential therapeutic targets for novel drugs in digestive systerm cancer (Figure 2). For example, overexpression of ciRS-7 could inhibit miR-7-mediated tumor suppression via antagonizing miR-7-mediated PTEN/PI3K/Akt pathway in gastric cancer [73] and EGFR/RAF1/MAPK pathway in colorectal cancer [79]. Furthermore, the expression of circRNA is regulated by Pol II promoters, hence disease-activated control elements can be used to limit circRNA expression in malignant cells, while cell-specific promoters can be used to control circRNA expression in certain cell types [90]. Gene knockout is another classical approach to cause circRNA function deficiency, thus using this technology may remove the carcinogenic circRNA. Rajewsky et al. have successfully created an in vivo loss-of-function model from the mouse genome by CRISPR/Cas9 system targeting Cdr1as [33]. It offers an unprecedented opportunity for removing oncogenic circRNA. Another example is circLARP4. It can upregulate the expression of large tumor suppressor kinase 1 (LATS1) by sponging miR-424 and AGO2 protein. Then the LATS1 downstream effectors were downregulated to suppress gastric tumorigenesis and progression [75]. This unique cellular capacity of circRNA to sponge miRNA and proteins also makes it a promising therapeutic vector. Exogenous introduction of synthetic circRNA with multiple binding sites for specific oncogenic miRNA and/or proteins, namely artificial sponges, may restore the normal regulatory network to control proliferation or induce apoptosis in cancer cells [34]. Moreover, circRNA sponge will show more potent anti-cancer effect than linear miRNA sponge [91] due to its potentially low off-target effect as well as its specific and stable structure. Ultimately, since circRNAs can modulate the stability of some proteins like p53 [92] and STAT3 [93] through circRNA-miRNA-mRNA axis to participate in antitumor immunity, circRNAs may be a promising target for tumor immunotherapy [94]. In addition, because of the stable and specific properties of circRNAs, they may serve as tumor antigens for immune response [94-96]. For example, purified circRNAs can activate innate immunity and be recognized by RIG-I. Therefore, foreign circRNAs from tumor cells may also activate immunocytes to fight tumors.
Figure 2.
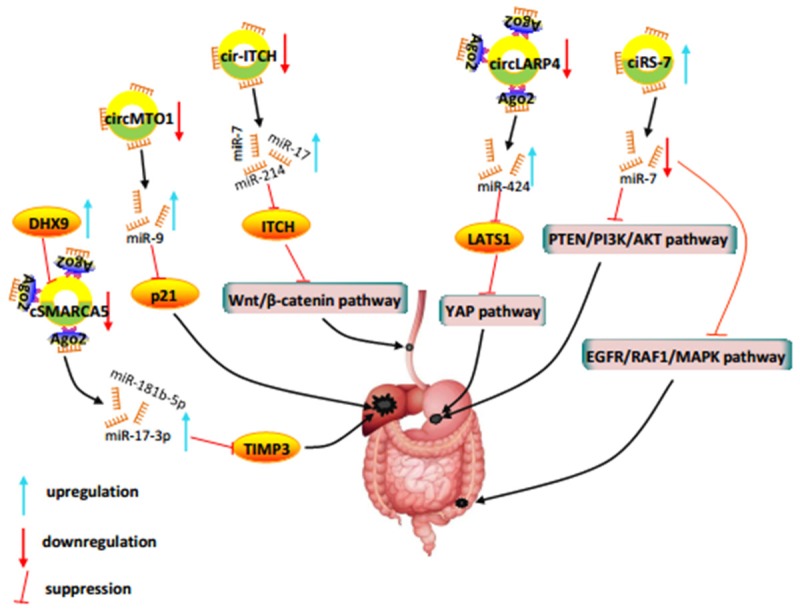
CircRNA-mediated mechanisms in digestive system cancer. CircRNA functions as the sponge of miRNAs. The expression dysregulation of circRNA will affect the expression of related miRNA target genes via upregulation or downregulation of free miRNA, ultimately giving rise to cancer.
Future perspectives
CircRNAs were previously thought to be junk by-products in the process of gene transcription [97,98]. However, accumulating researches in different cell types and tissues were performed to explore underlying functions of circRNAs. CircRNAs were confirmed to accomplish a remarkable multitude of biological functions such as regulating transcription or splicing, serving as miRNA sponges, interacting with RBPs, translating proteins, and so on. They are also involved in the biogenesis and development of many diseases, especially in cancer. Furthermore, some tools have been developed to annotate circRNAs [99,100], such as circRNA_finder [101], CIRCexplorer [35], find_circ [6], UROBORUS [102], KNIFE [103], CIRI [104], MapSplice [105], segemehl [106], and NCLscan [107]. These tools help to expand our understanding of the complex information regarding circRNAs in eukaryotic transcriptomes. As well, these tools were suggested to be used combinedly to improve the sensitivity and specificity of circRNA identification. Despite such useful state-of-the-art circRNA detection tools exist, there is still a lack of a standardized naming system for circRNA research.
More remarkably, chromosomal translocation sometimes generates fusion genes with oncogenic potential [108]. The phenomenon has been described in many hematological and solid malignancies. In addition to creating coding fusion mRNAs, cancer-associated chromosomal translocation could result in aberrant formation of circular RNAs, namely fusion circRNAs (f-circRNA) [109,110]. Moreover, f-circRNAs, with tumor-promoting properties, promote cell viability and resistance upon therapy, and contribute to cellular transformation. Nevertheless, there is still limited knowledge about the underlying mechanisms of f-circRNA to drive tumorigenesis and it still needs to be elucidated in more details.
Taken together, these studies expand the current knowledge and add complexity regarding molecular mechanisms involved in cancer occurrence and progression. New candidate cancer-associated circRNA genes are being identified and their molecular mechanisms are being elucidated. CircRNAs offer new possibilities for understanding cancer pathogenesis. As previously reviewed, many circRNAs have been verified to be potential markers of diagnosis, progression, or prognosis in digestive system cancer (Figure 3). Besides, large numbers of novel circRNAs are likely to be identified in the near future. An advanced understanding for circRNA in digestive system cancer will undoubtedly provide beneficial insights and generate new hypothesis regarding disease pathogenesis, which might eventually take breakthroughs in clinical application.
Figure 3.
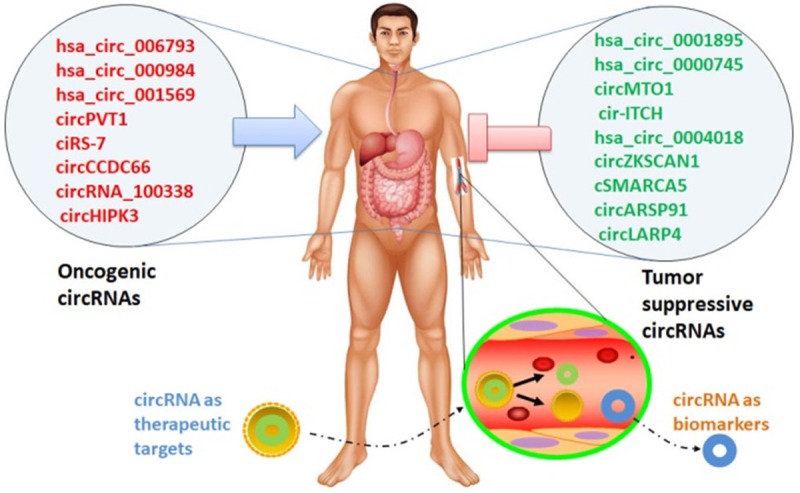
Cancer-promoting and cancer-suppressing circRNAs in digestive system cancers. Emerging evidences have demonstrated that circRNA expression is dysregulated in digestive system cancers. Different circRNAs play different roles, and they are antagonistic to each other. Some circRNAs may play oncogenic roles, whereas others play tumor suppressive roles in cancer. Moreover, circRNAs may serve as potential therapeutic targets for novel drugs and potential biomarkers for diagnosis and prognosis.
Acknowledgements
This work was supported by State Key Laboratory of Cancer Biology (Fourth Military Medical University), No. CBSKL201720; Natural Science Foundation of Hubei Province, No. 2017CFB457; National Natural Science Foundation of China, No. 81372663 and No. 81772969.
Disclosure of conflict of interest
None.
References
- 1.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 3.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, Dudekula DB, Abdelmohsen K, Gorospe M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 8.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Wu H, Wang Y, Zhu S, Liu J, Fang X, Chen H. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J Dairy Sci. 2016;99:4750–60. doi: 10.3168/jds.2015-10381. [DOI] [PubMed] [Google Scholar]
- 10.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–11. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–20. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, Wu F, Chao J, Liu P, Hu G, Zhang JH, Yao H. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, Lu L, Caffarelli E, Shneider NA, Morlando M, Bozzoni I. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Zhang L, Zhang K, Zhang G, Hu Y, Sun X, Zhao C, Li H, Li YM, Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis. 2018;77:770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, Jiang R, Yang X, Guo H, Fang S, Zhang Y, Cheng Y, Wang J, Yao H, Chao J. circRNA Mediates Silica-induced macrophage activation Via HECTD1/ZC3H12A-dependent ubiquitination. Theranostics. 2018;8:575–92. doi: 10.7150/thno.21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, Liu B, Li XM, Yao J, Jiang Q, Yan B. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–77. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 22.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–50. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–61. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–19. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y, Ao Y. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223–30. doi: 10.1016/j.omtn.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 29.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–65. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297–325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 33.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017:357. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 34.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 38.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 39.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 40.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–31. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 41.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18:780–8. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 43.Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–91. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–9. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 46.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatomer DC, Wilusz JE. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol Cell. 2017;66:1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–7. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 50.AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A. 2014;111:14542–7. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–9. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018:110. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of CircRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL, Zhou H, Yang JH, Qu LH. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196–202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–92. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 62.Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, Wei L, Han L, He C. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng X, Chen Q, Zhang P, Chen M. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics. 2017;33:3314–6. doi: 10.1093/bioinformatics/btx446. [DOI] [PubMed] [Google Scholar]
- 64.Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–15. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, Jin X, Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun J, Yuan X, Li X, Wang D, Shan T, Wang W, Wan Q, Wang X, Yan J, Gao S. Comparative transcriptome analysis of the global circular RNAs expression profiles between SHEE and SHEEC cell lines. Am J Transl Res. 2017;9:5169–5179. [PMC free article] [PubMed] [Google Scholar]
- 71.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 72.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP CONCORD Working Group. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of Circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 74.Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, Guo J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317699125. doi: 10.1177/1010428317699125. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Li J, Yu J, Liu H, Shen Z, Ye G, Mou T, Qi X, Li G. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 2017;8:22936–22943. doi: 10.18632/oncotarget.15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ, Wu WD. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674–83. doi: 10.18632/oncotarget.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 84.Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res. 2017;16:3891–3902. doi: 10.1021/acs.jproteome.7b00519. [DOI] [PubMed] [Google Scholar]
- 85.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, Liu B, Yang Y. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, Sun SH, Yang F, Zhou WP. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 88.Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, Zhou J, Tang ZY, Huang XY. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, Lin H, Liang X, Cai X. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171. doi: 10.1038/cddis.2017.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2017;37:555–65. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W, Zhang X, Su J, Chen X, Chen Q, Shou P, Xu C, Du L, Lin L, Xie N, Zhang L, Wang Y, Shi Y. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene. 2014;33:3830–3838. doi: 10.1038/onc.2013.355. [DOI] [PubMed] [Google Scholar]
- 93.Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu , Gupta S, Yang W, Yang BB. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol. 2018;9:9. doi: 10.3389/fimmu.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cadena C, Hur S. Antiviral immunity and circular RNA: no end in sight. Mol Cell. 2017;67:163–164. doi: 10.1016/j.molcel.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 99.Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13:e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, MacLeod JN, Chiang DY, Prins JF, Liu J. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt LM, Teupser D, Hackermuller J, Stadler PF. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016;44:e29. doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–54. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oncogenic circular RNAs arise from chromosomal translocations. Cancer Discov. 2016;6:OF20. doi: 10.1158/2159-8290.CD-RW2016-068. [DOI] [PubMed] [Google Scholar]
- 110.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 111.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S, Wang Z, Wen P, Yang H, Shi X, Pan J, Ye H. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169–48177. doi: 10.18632/oncotarget.18327. [DOI] [PMC free article] [PubMed] [Google Scholar]


