Abstract
Ovarian cancer has the highest mortality rate of all gynecologic malignancies. Identification of new biomarkers is highly needed due to its late diagnosis and high recurrence rate. The objective of this study was to identify mechanisms of therapy resistance and potential biomarkers by analyzing mRNA and protein expression from samples derived from patients with platinum-sensitive and -resistant ovarian cancer (total cohort n = 53). The data revealed new candidates for targeted therapies, such as GREB1 and ROR2. We showed that the development of platinum resistance correlated with upregulation of ROR2, whereas GREB1 was downregulated. Moreover, we demonstrated that high levels of ROR2 in platinum-resistant samples were associated with upregulation of Wnt5a, STAT3 and NF-kB levels, suggesting that a crosstalk between the non-canonical Wnt5a-ROR2 and STAT3/NF-kB signaling pathways. Upregulation of ROR2, Wnt5a, STAT3 and NF-kB was further detected in a platinum-resistant cell-line model. The results of the present study provided insight into molecular mechanisms associated with platinum resistance that could be further investigated to improve treatment strategies in this clinically challenging gynecological cancer.
Introduction
Epithelial ovarian cancer (EOC) accounts for the majority of mortality from gynecological cancers, with diagnosis often at a late stage. Currently, the golden standard of treatment is primary debulking surgery (PDS) followed by platinum-based chemotherapy [1]. Although most patients initially respond to chemotherapy, cancer cells will eventually develop resistance leading to relapse [2]. Despite intensive efforts to improve targeted therapy in EOC, the five-year survival rate is still only 30% for advanced disease [3]. Therefore, increased knowledge about mechanisms of platinum resistance in EOC treatment is needed in search for cure.
Genetically complex and unstable high-grade serous ovarian cancer subtype (HGSC), accounting for approximately 50–70% of EOC, represents the most aggressive histological subtype [4]. A large-scale integrated genomic data analysis for HGSC identified TP53 mutations in almost 96% of tumors. Recurrent somatic mutations were found in nine other genes including NF1, BRCA1, BRCA2, RB1 and CDK12, as well as DNA copy number aberrations and promoter methylation events, indicating biological and molecular heterogeneity that should be considered when developing novel therapeutic strategies [5].
While only 10%–15% of ovarian cancer patients carry BRCA1 or BRCA2 mutations in their germline, ∼50% of ovarian cancers exhibit a defect in the homologous recombination (HR) repair of DNA [6]. PARP-1 enzyme became an attractive target for chemotherapeutics for its crucial function in single-strand breaks (SSBs) DNA repair mechanism through base excision repair (BER) pathway [7]. The concept of synthetic lethality has been used in genetic studies to determine functional interactions and compensation among genes for decades and has also been exploited in the development of PARP (Poly (ADP-ribose) polymerase) inhibitors [8]. The responsiveness to platinum and PARP inhibitors associates with so called BRCAness profile showing independent prognostic value [9], [10].
The chemotherapy resistance can arise due to multiple mechanisms, such as drug target alteration, re-activation or amplification of the oncogenic pathway, activation of parallel pathways, increased DNA damage tolerance/repair, and deregulation of growth factor receptors among others [11]. Deregulation of apoptosis and altered phosphorylation (intracellular signaling), as well as metabolic pathways represent the two main biological processes responsible for oncogene-mediated drug resistance in ovarian cancer [12]. In this context, activation of PI3K/AKT cell survival pathway plays a pivotal role with NF-kB and STAT3 as the main mediations of these intracellular events. On the other hand, tumor suppressor genes such as BRCA1, BRCA2, MLH1 and p21 contribute to ovarian cancer drug resistance via alterations in the DNA damage and repair mechanisms, whereas RASSF1, TP53 and TP73 impair the apoptotic machinery for the same outcome [13]. Epithelial-to-mesenchymal transition (EMT) has also been implicated in HGSC invasiveness and chemoresistance, and in vitro studies using ovarian cancer cell lines have shown that more aggressive, mesenchymal-type cells are more resistant to cisplatin treatment [14]. An important signaling cascade involved in EMT is the Wnt signaling, with increasing evidence suggesting that β-catenin-independent pathway via Wnt5a/ROR1/ROR2 has a critical role in EMT and chemoresistance [15], [16], [17], [18]. Consequently, therapies targeting these pathways may offer means to overcome drug resistance.
Active and also productive research in the field of cancer therapy has led to an improved understanding of the molecular mechanisms, providing insight into the development of cancer. This new data has led to the development of new treatment options for cancer patients, including targeted therapies and associated biomarker tests that can select which patients are most likely to respond [19]. The aim of this study was to identify candidate genes and their molecular pathways involved in the pathogenesis of ovarian cancer associating with platinum resistance. Overcoming the paucity of obtaining large collection of tumor samples available for molecular profiling, we investigated differences in mRNA and protein expression between ovarian cancer samples derived from two clinically and molecularly distinct patient cohorts namely high PARP/platinum-sensitive and low PARP/platinum-resistant HGSC cohorts. This comparison aimed at distinction of two cohorts with extremely different clinical behavior. Finally, this analysis led to identification of GREB1 and ROR2 that showed significant differential expression profile between the two groups. Our data suggest new predictive biomarkers for ovarian cancer drug resistance development warranting further investigations.
Materials and Methods
Study Cohort and Tissue Samples
The study was carried out at the University of Tampere and Tampere University Hospital (TAUH), Tampere, Finland. The study protocol was approved by the Ethics Committee of TAUH (identification code ETL-R11137).
The microarray study cohort consisted of 12 HGSC patients who participated in a prospective study addressing PARP enzyme activity in fresh ovarian cancer tumor samples [20]. The selection of this patient subcohort was based on PARP values (high PARP/low PARP, cut off 203 pg/ml, which corresponded to the median value of PARP) and platinum sensitivity/resistance (treatment response), with no differences in respect of age and FIGO (International Federation of Gynecology). Platinum sensitivity was defined as no recurrence within 12 months after the completion of first-line platinum-based chemotherapy. Patient characteristics are presented in Table 1.
Table 1.
Characteristics of the Study Patients in the Microarray Cohort (n = 12)
| Characteristic | Platinum Sensitive* n (%) | Platinum Resistant* n (%) |
|---|---|---|
| All | 6 | 6 |
| PDS† | 5 | 2 |
| NACT‡ | 1 | 4 |
| PARP§ low | 0 | 6 |
| PARP§ high | 6 | 0 |
| Age | ||
| mean (SD) | 65 | 62 |
| median (range) | 63 (46–78) | 62 (55–79) |
| Grade 3¶ | 6 (100%) | 6 (100%) |
| Stage# | ||
| FIGO st I | 0 | 0 |
| FIGO st II | 0 | 0 |
| FIGO st III and IV | 6 | 6 |
| Histology | ||
| serous | 6 (100%) | 6 (100%) |
| PFS** (months) | 28 | 3.5 |
Sensitivity defined as relapse or event-free interval> 12 months after completion of platinum based 1st line therapy.
PDS - primary debulking surgery.
NACT - neoadjuvant therapy.
PARP - PARP activity in fresh frozen tumor tissue was assessed by an enzymatic chemiluminescense assay in a previous study [20]; the cut-off level for high PARP activity was set to 203 pg/ml corresponding to median value.
Grade – Grade 3 represents high grade tumors.
FIGO - International Federation of Gynecology.
PFS - progression free survival.
The validation cohort of the microarray data consisted of all the patients (n = 53) that participated in the previous prospective study [20]. The median follow-up time of patients was 31 months. The validation cohort is described in Table 2.
Table 2.
Characteristics of the Study Patients in the Validation Cohort (n = 53)
| Characteristic | |
|---|---|
| Patients in study, n | 53 |
| Age at surgery, yrs. (SD) | 66 (9.3) |
| BMI, mean (SD) | 26.8 (6.2) |
| Median follow-up, months (range) | 31 (2–50) |
| Ca 125 level (kU/L) before treatment, median (range) | 523 (30–4728) |
| FIGO* Stage, n (%) | |
| Stage 1 | 2 (3,7%) |
| Stage 2 | 3 (5,6%) |
| Stage 3 | 34 (64,3%) |
| Stage 4 | 14 (26,4%) |
| Histology, n (%) | |
| Serous | 46 (86,8) |
| Endometroid | 4 (7,6%) |
| Papillar | 2 (3,7%) |
| Mucinous | 0 (0%) |
| Carcinosarcoma | 0 (0%) |
| Transitional cell | 1 (1,9%) |
| Grade†, n (%) | |
| Grade 1 and 2 | 10 (18%) |
| Grade 3 | 43 (82%) |
| Sensitivity to platinum therapy‡, n (%) | |
| Sensitive | 25 (47,2%) |
| Resistant | 15 (28,3%) |
| Partial sensitive | 13 (24,5%) |
| Neoadjuvant therapy, n (%) | 19 (36%) |
| Recurrence | 36 (68%) |
| Death | 22 (42%) |
FIGO = International Federation of Gynecology.
Grade – Grade 3 represents high grade tumors, Grade 1 and 2 low grade tumors.
Sensitive: Recurrence >12 months after completion of platinum-based 1st line therapy; Resistant: Recurrence ≤6 months after completion of platinum-based 1st line therapy; Partially sensitive: Recurrence 6–12 months after completion of platinum-based 1st line therapy.
For further investigation of ROR2 in EOC, a retrospective subcohort was chosen from the study cohorts described above consisting of a subgroup of patients who had not received NACT (neoadjuvant chemotherapy) and were divided in two categories based on treatment response, i.e. either platinum-sensitive or platinum-resistant (Table 3).
Table 3.
Characteristics of the study patients in the ROR investigation subcohort (n = 30)
| Characteristic | |
|---|---|
| Patients in study, n | 30 |
| Age at surgery, yrs mean (SD) | 66 (9.3) |
| Median follow-up, months (range) | 31 (2–50) |
| FIGO* Stage, n (%) | |
| Stage 1 | 2 (6,7%) |
| Stage 2 | 4 (14%) |
| Stage 3 | 17 (56%) |
| Stage 4 | 7(23,3%) |
| Histology, n (%) | |
| Serous | 22(73,3%) |
| Endometroid | 2 (6,7%) |
| Papillar | 2 (6,7%) |
| Mucinous | 2 (6,7%) |
| Carcinosarcoma | 1 (3,3%) |
| Transitional cell | 1 (3,3%) |
| Grade†, n (%) | |
| Grade 1 and 2 | 8 (26,7%) |
| Grade 3 | 22 (73,3%) |
| Response to platinum therapy‡, n (%) | |
| Sensitive | 20 (66,7%) |
| Resistant | 10 (33,3%) |
| Recurrence | 20 (66,7%) |
| Death | 9 (30%) |
FIGO = International Federation of Gynecology.
Grade – Grade 3 represents high grade tumors, Grade 1 and 2 low grade tumors.
Sensitivity defined as relapse or event-free follow up time> 12 months after completion of platinum based 1st line therapy.
The tumor tissue samples were collected at surgery, two samples approximately 0.5 cm were chosen at the operation room from macroscopically visible tumor and were snap-frozen with liquid nitrogen and stored in −70 °C. The findings from the corresponding archival surgical tumor specimens were assessed by experienced pathologists as part of routine diagnostics at the Department of Pathology at TAUH.
mRNA Microarray
Total RNA from ovarian cancer fresh frozen samples was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. The quality and integrity of the RNA was assessed using Fragment Analyzer parallel capillary electrophoresis (Advanced Analytical Technologies, Ankeny, IA, USA). The RNA was subsequently labeled and hybridized using the Agilent gene expression microarray kit (Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer's instruction. Briefly, the RNA from the clinical samples was labeled with Cy3 fluorochrome and subsequently co-hybridized with Cy5 labeled Xpress Ref TM Human Universal Reference Total RNA (SuperArray Bio-science Corporation) as a control. The hybridization was carried out for 21 hours on Agilent 4X44K human gene expression array slides. The slides were subsequently scanned on an Agilent C scanner and raw data were extracted using the Agilent Feature Extraction software ver. 11.0.1.1 and quantile normalized. A fold-change cutoff of 2 was used to determine differential mRNA expression as well as q-value and signal intensity.
qRT-PCR
Putatively differentially expressed genes (ROR2, CAST, ATP6V1D, GUCY1A3, TMOD1, MYCN, DLK1, PLEKHG4B, GREB1, B4GALNT4, SLC35F3, PTCH2, TNNC1, BNC1) were selected from the array results based on fold change, q-value, signal intensity and literature data and were validated by quantitative real-time polymerase chain reaction (qRT-PCR). Total RNA from ovarian cancer fresh frozen tumor samples were extracted with TRIzol as described above and were reverse transcribed using random hexamere primers and MultiScribe reverse transcriptase (Thermo Fischer Scientific, Waltham, MA, USA). Quantitative Real Time PCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fischer Scientific) on a BioRad CFX96 ™ Real-Time PCR Detection System (BioRad Laboratories, Hercules, CA, USA). Each sample was run in duplicate and expression values were normalized against the TATA-binding protein (TBP). The primer sequences for the two validated genes are as follows:
| GREB1 fw | 5’ATGGGAAATTCTTACGCTGGAC |
|---|---|
| GREB1 rev | 5’CACTCGGCTACCACCTTCT |
| ROR2 fw | 5’GTGCGGTGGCTAAAGAATGAT |
| ROR2 rev | 5’ATTCGCAGTCGTGAACCATATT |
| TBP fw | 5’GAATATAATCCCAAGCGGTTTG |
| TBP rev | 5’ACTTCACATCACAGCTCCCC |
Western blotting
Frozen tumor pieces were thawed, washed 2X with cold PBS and pestered to lyse in lysis buffer (50 mM Tris–HCl pH 7.5, 10% glycerol, 150 mM NaCl, 1 mM EDTA, 1% Triton-x-100, 50 mM NaF) supplemented with protease and phosphatase inhibitor cocktails (Bimake, Houston, TX, USA). Lysates were mixed with 4X Laemmli loading buffer and subjected to SDS-PAGE gel electrophoresis and Western blotting. The primary antibodies used were as following: pAkt S473 (#4060), pMEK1/2 S217/221 (#9121), NF-κB p65 (#6956), pPI3K p85 Y458/p55 Y199 (#4228), PI3K p85α (#13666), Rac-1 (#4651), pSTAT3 Y705 (#9145), STAT3 (#9139), Wnt5a/b (#2530) (Cell Signaling Technology, Danvers, MA, USA); Akt (#sc-5298), Bcl-2 (#sc-7382), MEK1/2 (#sc-6250), β-tubulin (#sc-166,729) (Santa Cruz, Dallas, TX, USA); ROR1 6D4 (Dr. Riddel lab, ref. Balakrishana et al. 2016); ROR2 (#565550, BD Biosciences, San Jose, CA, USA). Secondary antibodies: IRDye® 800CW Donkey anti-Mouse IgG, or IRDye® 680RD Donkey anti-Rabbit IgG (LI-COR, Lincoln, NE, USA). Blots were scanned and quantified using Odyssey CLx and Image Studio software (LI-COR). Protein quantification was normalized to β-tubulin expression level for each sample.
Cell Culture
A2780 and A2780cis cells were purchased from Merck & Company Inc. (Kenilworth, NJ, USA) and cultured according to manufacturer's recommendations. The A2780 line was maintained in medium containing 1 μM cisplatin (Selleckchem, Munich, Germany). The cisplatin EC50 response of A2780 and A2780cis cells was validated by incubating cells with increasing concentration of cisplatin for 3 days and cell viability was determined using CellTiterGlo (CTG) Assay (Promega, USA) according to manufacturer's instructions.
qRT-PCR of the Ovarian Cancer Cell Lines
RNA was collected from A2780 and A2780cis using TRI Reagent® (Molecular Research Center Inc. Cincinnati, OH, USA) according to the manufacturer's protocol. qRT-PCR was performed as described for the ovarian cancer clinical samples. Each cell line was run in 4 replicates and the expression of ROR2 and GREB1 was normalized against TBP.
Statistical Analysis
The statistical analysis of mRNA microarray data was implemented in R using packages limma and preprocessCore of Bioconductor project [21], [22], [23]. Data was quantile normalized and probe sets were summarized by choosing the probes with the highest average expression [22]. Using limma approach, differential expression was identified between patients who had low PARP value and were platinum resistant, and patients who had high PARP value and were platinum sensitive [23]. P-values were obtained by the empirical Bayes moderated t-test. A fold-change cutoff of 2 was used to determine differential mRNA expression. The results were interpreted by principal component analysis [24] and hierarchical clustering.
qRT-PCR data were analyzed using the Student's t-test and Mann–Whitney U test where appropriate. The Kaplan–Meier regression analyses were used to estimate the survival rates from the date of surgery (primary debulked patients) or from the date of the first dose of neoadjuvant therapy until the date of the event of interest. For progression-free survival (PFS), the event of interest was a recurrence or death, whichever occurred first. Patients alive at the last follow-up without a recurrence were censored at the last follow-up date. Statistical analysis was performed using GraphPad Prism 6 software for Windows (GraphPad Software Inc., La Jolla, CA, USA). A p-value less than 0.05 was considered significant.
Oncomine (https://www.oncomine.org/resource/login.html) and Kaplan–Meier plotter (http://kmplot.com/analysis/index.php?p=service&cancer=ovar) databases were searched for gene expression and survival data.
Results
Microarray Analysis of HGSC Patient Samples
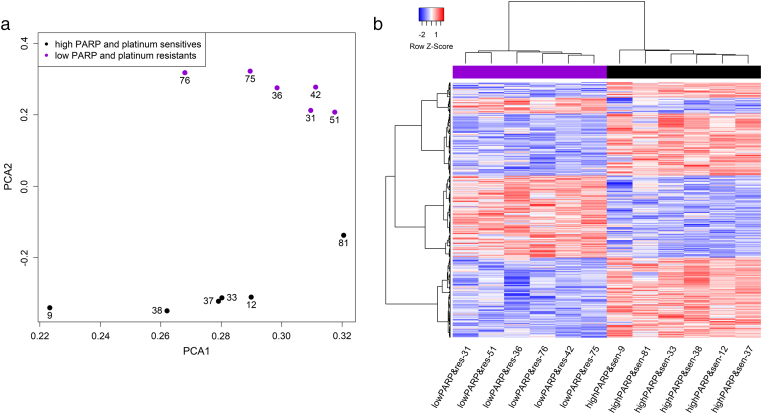
In order to identify differentially expressed genes between ovarian cancer samples with platinum-sensitivity with high PARP levels and samples with platinum resistance with low PARP levels (n = 12), gene expression microarray was performed on total RNA isolated from the freshly frozen tumor samples. The analysis of gene expression showed a total of 3001 differentially expressed genes between the two comparison groups when a log fold change cutoff 2 was implemented. In this comparison, 1463 genes were downregulated and 1538 genes were upregulated. 50 most upregulated and 50 most downregulated genes are summarized in Table 4, Table 5, respectively. The comparison of patient groups according to PARP levels and treatment responses are shown in Figure 1, a-b.
Table 4.
The 50 Most Upregulated mRNAs in High PARP and Platinum Sensitive OC Patient Samples in Comparison to low PARP and Platinum Resistant Samples.
| Gene name | Gene description | log2-fold change | p-value | q-value |
|---|---|---|---|---|
| COLEC11 | collectin subfamily member 11 | 3.68E+ 00 | 1.29E-02 | 0.2588936 |
| MYCN | MYCN proto-oncogene. bHLH transcription factor | 3.61E+ 00 | 2.74E-04 | 0.1050305 |
| ESM1 | endothelial cell specific molecule 1 | 3.18E+ 00 | 1.64E-04 | 0.1028588 |
| IGF1R | insulin like growth factor 1 receptor | 3.08E+ 00 | 5.29E-03 | 0.211962 |
| TNNC1 | troponin C1. slow skeletal and cardiac type | 3.04E+ 00 | 1.25E-05 | 0.0472319 |
| LGSN | lengsin. Lens protein with glutamine synthetase domain | 3.02E+ 00 | 1.25E-02 | 0.2588936 |
| LOC100134423 | uncharacterized LOC100134423 | 2.97E+ 00 | 2.71E-04 | 0.1050305 |
| DLK1 | delta like non-canonical Notch ligand 1 | 2.89E+ 00 | 4.17E-03 | 0.2013983 |
| CRYGC | crystallin gamma C | 2.86E+ 00 | 2.37E-03 | 0.1822358 |
| A_33_P3286709 | NA | 2.82E+ 00 | 9.15E-05 | 0.0864965 |
| PLEKHG4B | pleckstrin homology and RhoGEF domain containing G4B | 2.82E+ 00 | 1.17E-03 | 0.1525447 |
| TSPAN8 | tetraspanin 8 | 2.78E+ 00 | 2.99E-02 | 0.3064189 |
| ENPP6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | 2.73E+ 00 | 1.20E-02 | 0.2563874 |
| FLJ30901 | uncharacterized protein FLJ30901 | 2.60E+ 00 | 3.26E-04 | 0.1050305 |
| PROM1 | prominin 1 | 2.59E+ 00 | 3.02E-02 | 0.3079072 |
| COL22A1 | collagen type XXII alpha 1 chain | 2.58E+ 00 | 2.64E-03 | 0.1844765 |
| KIAA1324 | KIAA1324 | 2.58E+ 00 | 5.48E-04 | 0.1261184 |
| PTCH2 | patched 2 | 2.57E+ 00 | 1.93E-05 | 0.0472319 |
| SLC35F3 | solute carrier family 35 member F3 | 2.54E+ 00 | 3.87E-05 | 0.0596903 |
| GABRG3 | gamma-aminobutyric acid type A receptor gamma3 subunit | 2.51E+ 00 | 4.50E-05 | 0.0628517 |
| LOC613266 | uncharacterized LOC613266 | 2.50E+ 00 | 4.89E-02 | 0.3485897 |
| LCE1E | late cornified envelope 1E | 2.49E+ 00 | 2.28E-04 | 0.1050305 |
| B4GALNT4 | beta-1.4-N-acetyl-galactosaminyltransferase 4 | 2.49E+ 00 | 1.24E-05 | 0.0472319 |
| STC2 | stanniocalcin 2 | 2.48E+ 00 | 2.86E-04 | 0.1050305 |
| FOXL2NB | FOXL2 neighbor | 2.48E+ 00 | 1.04E-01 | 0.4447586 |
| AK124496 | NA | 2.47E+ 00 | 2.78E-02 | 0.3020977 |
| CU677518 | NA | 2.47E+ 00 | 2.82E-06 | 0.0275612 |
| NM_130777 | NA | 2.45E+ 00 | 1.10E-02 | 0.2525629 |
| NR_102701 | NA | 2.43E+ 00 | 2.11E-03 | 0.1762016 |
| NSG1 | neuronal vesicle trafficking associated 1 | 2.43E+ 00 | 1.05E-03 | 0.1525447 |
| STAR | steroidogenic acute regulatory protein | 2.41E+ 00 | 5.08E-03 | 0.2110634 |
| MCTS2P | malignant T-cell amplified sequence 2. pseudogene | 2.40E+ 00 | 5.43E-03 | 0.2134259 |
| TRABD2A | TraB domain containing 2A | 2.36E+ 00 | 4.89E-03 | 0.2078187 |
| DEPTOR | DEP domain containing MTOR interacting protein | 2.35E+ 00 | 4.70E-04 | 0.125416 |
| CLEC4GP1 | C-type lectin domain family 4 member G pseudogene 1 | 2.35E+ 00 | 4.56E-03 | 0.2021535 |
| LINC01405 | long intergenic non-protein coding RNA 1405 | 2.33E+ 00 | 6.95E-04 | 0.1391319 |
| COL2A1 | collagen type II alpha 1 chain | 2.32E+ 00 | 1.82E-02 | 0.2816697 |
| HS6ST2 | heparan sulfate 6-O-sulfotransferase 2 | 2.31E+ 00 | 2.15E-02 | 0.2871745 |
| PRAME | preferentially expressed antigen in melanoma | 2.30E+ 00 | 4.32E-05 | 0.0628517 |
| LINC02398 | long intergenic non-protein coding RNA 2398 | 2.29E+ 00 | 1.08E-05 | 0.0472319 |
| GJB7 | gap junction protein beta 7 | 2.29E+ 00 | 3.18E-02 | 0.3122712 |
| PLCXD3 | phosphatidylinositol specific phospholipase C X domain containing 3 | 2.28E+ 00 | 6.03E-02 | 0.3707384 |
| ERVI-1 | endogenous retrovirus group I member 1 | 2.27E+ 00 | 1.28E-02 | 0.2588936 |
| ZNF556 | zinc finger protein 556 | 2.27E+ 00 | 1.46E-06 | 0.0213888 |
| NTS | neurotensin | 2.26E+ 00 | 6.56E-02 | 0.3828556 |
| BU963192 | NA | 2.25E+ 00 | 7.17E-05 | 0.0751488 |
| GMNC | geminin coiled-coil domain containing | 2.24E+ 00 | 1.52E-02 | 0.2689731 |
| A_33_P3274001 | NA | 2.24E+ 00 | 9.36E-05 | 0.0864965 |
| CRYGD | crystallin gamma D | 2.24E+ 00 | 2.58E-02 | 0.2964626 |
| GREB1 | growth regulation by estrogen in breast cancer 1 | 2.22E+ 00 | 1.22E-03 | 0.1542519 |
mRNAs selected for further validation are shown in bold.
Table 5.
The 50 Most Downregulated mRNAs in High PARP and Platinum Sensitive OC Patient Samples in Comparison to Low PARP and Platinum Resistant Samples
| Gene Name | Gene Description | Log2-Fold Change | p-Value | q-Value |
|---|---|---|---|---|
| EFEMP1 | EGF containing fibulin extracellular matrix protein 1 | −3.85E+ 00 | 1.50E-03 | 0.1587368 |
| FAP | fibroblast activation protein alpha | −3.66E+ 00 | 9.07E-04 | 0.1525447 |
| TMOD1 | tropomodulin 1 | −3.41E+ 00 | 2.92E-08 | 0.0008577 |
| BNC1 | basonuclin 1 | −3.36E+ 00 | 2.74E-04 | 0.1050305 |
| ENST00000453673 | NA | −3.22E+ 00 | 2.22E-02 | 0.2877726 |
| ENST00000411475 | NA | −3.18E+ 00 | 2.30E-02 | 0.2898232 |
| SCRG1 | stimulator of chondrogenesis 1 | −3.18E+ 00 | 1.64E-03 | 0.1627023 |
| PTGFR | prostaglandin F receptor | −3.04E+ 00 | 1.05E-03 | 0.1525447 |
| POSTN | periostin | −3.01E+ 00 | 1.10E-02 | 0.2525629 |
| ENST00000390237 | NA | −2.97E+ 00 | 1.54E-02 | 0.2695379 |
| BF175071 | NA | −2.92E+ 00 | 5.12E-02 | 0.3538495 |
| GUCY1A3 | guanylate cyclase 1 soluble subunit alpha | −2.89E+ 00 | 5.68E-05 | 0.0723946 |
| ENST00000390628 | NA | −2.87E+ 00 | 4.44E-03 | 0.2021325 |
| CXCL13 | C-X-C motif chemokine ligand 13 | −2.87E+ 00 | 1.02E-02 | 0.2487313 |
| ACKR4 | atypical chemokine receptor 4 | −2.86E+ 00 | 9.73E-05 | 0.0864965 |
| IGLL5 | immunoglobulin lambda like polypeptide 5 | −2.86E+ 00 | 3.46E-02 | 0.318167 |
| AREG | amphiregulin | −2.83E+ 00 | 1.43E-02 | 0.2670241 |
| A_33_P3251412 | NA | −2.81E+ 00 | 1.46E-02 | 0.2682963 |
| FYB1 | FYN binding protein 1 | −2.79E+ 00 | 2.51E-03 | 0.1843078 |
| CD38 | CD38 molecule | −2.78E+ 00 | 1.54E-03 | 0.1587368 |
| ENST00000468879 | NA | −2.77E+ 00 | 4.36E-02 | 0.3380632 |
| ENST00000390252 | NA | −2.73E+ 00 | 1.49E-02 | 0.2682963 |
| JCHAIN | joining chain of multimeric IgA and IgM | −2.70E+ 00 | 6.61E-03 | 0.2195342 |
| ENST00000390547 | NA | −2.69E+ 00 | 5.85E-03 | 0.2163421 |
| MEDAG | mesenteric estrogen dependent adipogenesis | −2.69E+ 00 | 4.49E-02 | 0.3403785 |
| CNR1 | cannabinoid receptor 1 | −2.68E+ 00 | 9.85E-03 | 0.2471112 |
| PRRX1 | paired related homeobox 1 | −2.68E+ 00 | 7.45E-04 | 0.1427284 |
| NR4A3 | nuclear receptor subfamily 4 group A member 3 | −2.67E+ 00 | 2.00E-02 | 0.2850432 |
| CH25H | cholesterol 25-hydroxylase | −2.65E+ 00 | 2.21E-03 | 0.1762175 |
| AB363267 | NA | −2.65E+ 00 | 4.23E-02 | 0.3347815 |
| ENST00000426099 | NA | −2.63E+ 00 | 4.11E-02 | 0.331398 |
| CCL18 | C-C motif chemokine ligand 18 | −2.57E+ 00 | 1.16E-04 | 0.0898927 |
| THBS2 | thrombospondin 2 | −2.57E+ 00 | 3.91E-02 | 0.327707 |
| ENST00000410078 | NA | −2.56E+ 00 | 5.30E-02 | 0.3580372 |
| BCHE | butyrylcholinesterase | −2.55E+ 00 | 1.00E-02 | 0.2471753 |
| ATP6V1D | ATPase H+ transporting V1 subunit D | −2.55E+ 00 | 1.54E-04 | 0.1028588 |
| ENST00000390323 | NA | −2.55E+ 00 | 2.42E-02 | 0.2937808 |
| CCR2 | C-C motif chemokine receptor 2 | −2.54E+ 00 | 9.37E-03 | 0.24523 |
| ENST00000479981 | NA | −2.54E+ 00 | 9.43E-03 | 0.24523 |
| CAST | calpastatin | −2.54E+ 00 | 1.02E-03 | 0.1525447 |
| ASPN | asporin | −2.52E+ 00 | 2.94E-02 | 0.3049408 |
| HBB | hemoglobin subunit beta | −2.52E+ 00 | 2.08E-02 | 0.2862875 |
| ENST00000390247 | NA | −2.51E+ 00 | 2.16E-02 | 0.2871745 |
| SULF1 | sulfatase 1 | −2.50E+ 00 | 1.80E-02 | 0.2805948 |
| EPYC | epiphycan | −2.49E+ 00 | 1.28E-02 | 0.2588936 |
| FGFBP1 | fibroblast growth factor binding protein 1 | −2.49E+ 00 | 1.03E-02 | 0.2499163 |
| LRRC15 | leucine rich repeat containing 15 | −2.48E+ 00 | 4.14E-02 | 0.3321649 |
| DOCK11 | dedicator of cytokinesis 11 | −2.45E+ 00 | 5.13E-04 | 0.1261184 |
| OOEP | oocyte expressed protein | −2.45E+ 00 | 4.23E-03 | 0.2013983 |
| ROR2 | receptor tyrosine kinase like orphan receptor 2 | −1.84E+ 00 | 2.83E-05 | 0.0506967 |
mRNAs selected for further validation are shown in bold.
Figure 1.
a. Scatterplot showing location of samples of high PARP and platinum sensitive and low PARP and platinum resistant in patients along the first two principal components (PC1 and PC2). Platinum sensitive and high PARP level samples are represented by black and low PARP and platinum resistant samples by purple dots.
b. Unsupervised clustering of the clinical samples shows differential expression patterns in patients with high PARP levels and platinum sensitivity compared to patients with low PARP levels and platinum resistant.
GREB1 and ROR2 are Significantly Differentially Expressed in Ovarian Cancer Tumor Samples Based on Platinum Sensitivity and PARP Levels
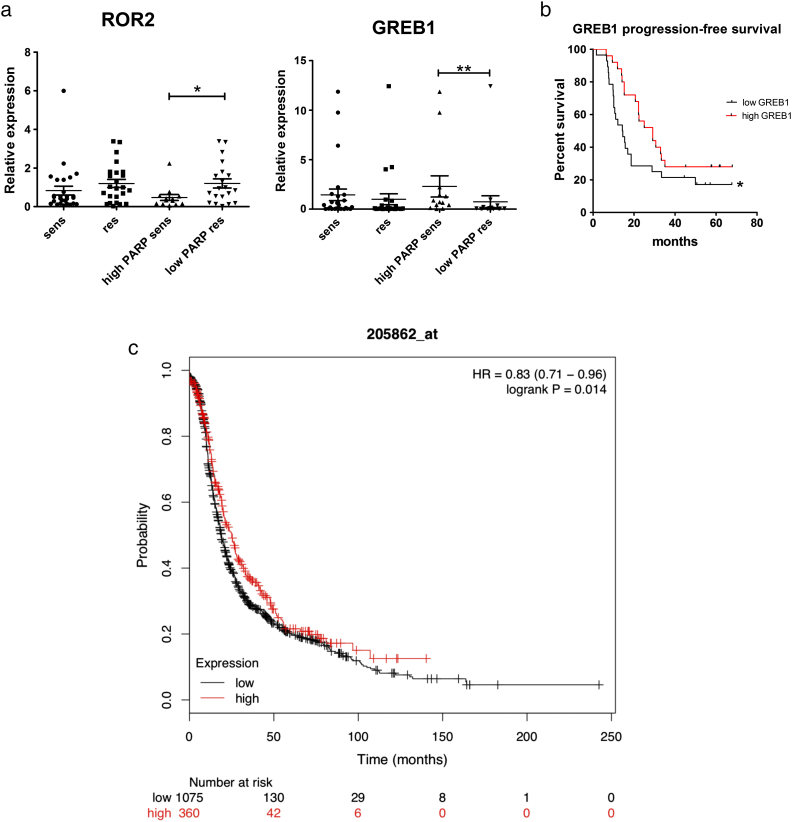
Fourteen differentially expressed mRNAs (namely: ROR2, CAST, ATP6V1D, GUCY1A3, TMOD1, MYCN, DLK1, PLEKHG4B, GREB1, B4GALNT4, SLC35F3, PTCH2, TNNC1, BNC1) from the microarray analysis were selected for validation by qRT-PCR in the cohort of 53 ovarian cancer patients, based on fold change, q-value, signal intensity and previous literature (Table 2). Two genes, namely ROR2 and GREB1, were significantly differentially expressed in high PARP/platinum-sensitive vs. low PARP/platinum-resistant groups (P = .02 and 0.002, respectively) (Figure 2, a-b). ROR2 was downregulated in microarray data in high PARP/platinum-sensitive tumor samples and the same result was obtained by qRT-PCR analysis in the validation cohort (n = 53). A trend towards statistically significant difference between the platinum sensitive and resistant groups regardless of PARP levels was observed (P = .058) (Figure 2a). The comparisons described above were based on treatment response as a significant clinical feature and issue of interest. In addition, previously determined PARP levels were taken into account to further investigate tumors from the BRCAness profile point of view. The platinum sensitive and high PARP level cohort is considered a group of better prognosis befitting with the BRCAness profile, whereas the platinum resistant and low PARP level a group with poorer prognosis.
Figure 2.
a. Expression of ROR2 and GREB1 in platinum sensitive (n = 38) vs resistant (n = 15) samples as well as high PARP level and platinum sensitive (n = 17) vs low PARP level and platinum resistant (n = 14) samples according to qRT-PCR (P = .02 and 0.002 respectively). The graphs show mean ± SEM.
b. Kaplan–Meier analysis of progression free survival (PFS) according to median level of GREB 1 concentration (log rank P = .019). Vertical lines represent censored patients.
c. Kaplan–Meier analysis of progression free survival (PFS) according to median level of GREB 1 concentration (log-rank P = .014) from Kaplan–Meier plotter database.
On the other hand, GREB1 was upregulated in microarray data in high PARP/platinum-sensitive tumor samples, and this result was validated by qRT-PCR (Figure 2b). Furthermore, GREB1 also showed a trend of overexpression in platinum sensitive samples, regardless of PARP level (P = .26).
GREB1 Expression Defines Platinum Sensitivity and Correlates with Longer PFS in Ovarian Cancer
Furthermore, we investigated whether GREB1 expression affects PFS in ovarian cancer patients (n = 53). High GREB1 expression was significantly associated with longer PFS as shown in Figure 2c (P = .019 in log-rank test). In Kaplan–Meier database search, which included 1465 ovarian cancer patients, a similar result was shown associating high GREB1 expression with better PFS (P = 0,014 in log-rank test), as shown in Figure 2d. In addition to Kaplan–Meier plotter database and Oncomine (website links are reported in statistical analysis section) were searched for the ROR2 and GREB1 genes. No similar results in expression correlation were found, however, no similar study settings were found (HGSC, treatment response comparison).
High ROR2 Expression in LowPARP/Platinum Resistant Ovarian Cancer Samples Correlated with Higher Wnt5a, STAT3 and NF-kB levels
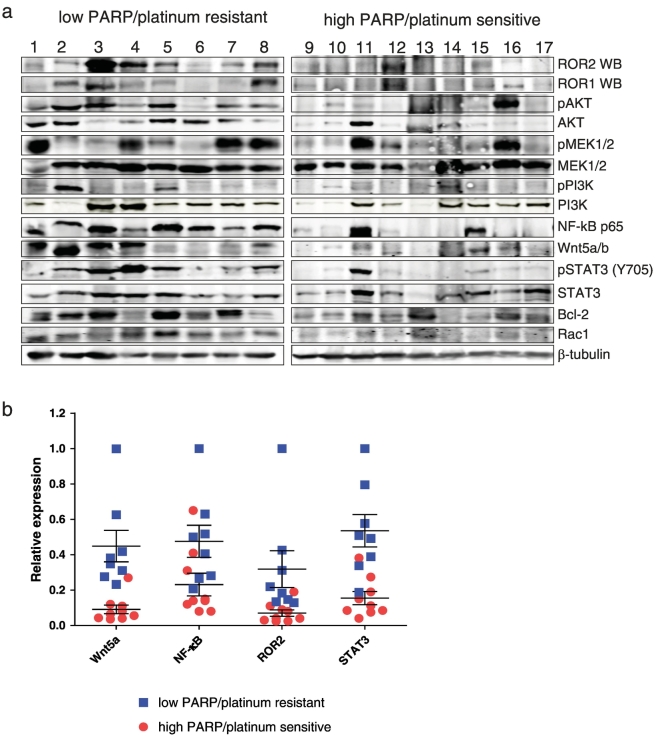
Previous data have shown no association with ROR2 expression and relapse-free survival [25] in ovarian cancer. However, ROR2 and ROR1 expression is increased in cisplatin resistant A2780 cell line compared to parental cells ([18]), and silencing their ligand Wnt5a in serous adenocarcinoma OVCAR3 cell line had greater effect in inhibiting cell migration and invasion than silencing either ROR alone [25]. Since ROR2 was significantly upregulated in low PARP/platinum-resistant patient samples in our microarray data, we decided to investigate the expression levels of Wnt5a, ROR1 and ROR2 proteins by Western blot in a subcohort of samples described in Table 3. As shown in Figure 3a-b, higher expression levels of Wnt5a, ROR1 and ROR2 proteins were found in low PARP/platinum-resistant group compared to high PARP/platinum-sensitive group. Moreover, upregulation of downstream signaling mediators such as pSTAT3 (Y705) and NF-kB was also observed in tumor lysates from low PARP/platinum resistant group. Noteworthy, patient samples with high levels of ROR1 and ROR2 from low PARP/platinum-resistant group showed higher pSTAT3 (Y705) levels (Figure 3a), indicating that STAT3 could mediate signaling downstream ROR1 and ROR2. Database search (Oncomine, Kaplan–Meier plotter; website links are reported in statistical analysis section) revealed no additional information regarding ROR in therapy response and ovarian cancer setting.
Figure 3.
Wnt5a/ROR2 expression is increased in lysates from platinum resistant tumors. A. Western blot analysis of lysates from platinum resistant vs. platinum sensitive patient samples with the indicated primary antibodies. B. Quantification of protein expression for Wnt5a, ROR2, STAT3 and NF-kB based on β-tubulin levels. Horizontal lines represent the average and errors bars are SEM (standard error of mean). After normalization, samples were quantified based on the highest expression level of all samples.
ROR2 and GREB1 Show Differential Expression in Cisplatin Resistant Ovarian Cancer Cell Line Model
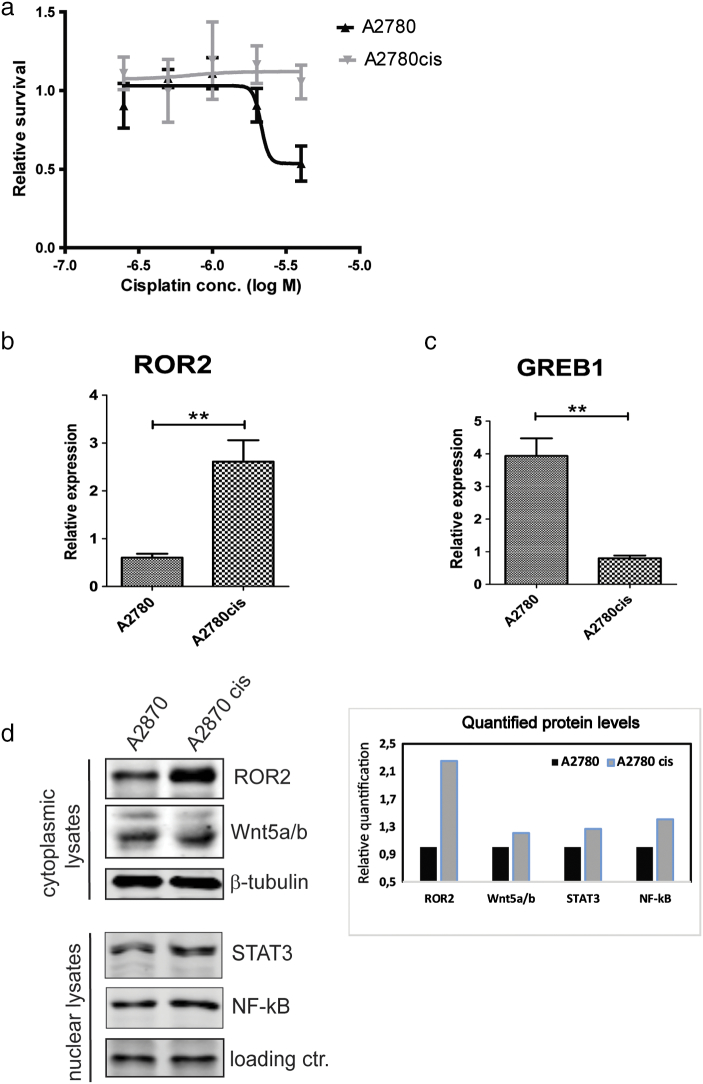
The human epithelial ovarian cancer cell line A2780 and its cisplatin resistant model A2780cis were selected to investigate the expression levels of ROR2 and GREB1. We confirmed the chemoresistant phenotype of A2780cis cells compared to A2780 parental cells by CTG assay and found significant increase of EC50 to cisplatin treatment for A2780cis cells (Figure 4a). The expression of both ROR2 and GREB1 mRNA level was investigated in A2780 and A2780cis cells (Figure 4b and c). We observed ROR2 mRNA upregulation in platinum resistant cell line A2780cis compared to platinum sensitive A2780 cells (P = .0046), as previously shown ([18]), whereas GREB1 mRNA level was downregulated in A2780cis compared to A2780 parental cells (P = .0012). Furthermore, A2780cis cells have increased protein expression levels of ROR2 and Wnt5a compared with the parental A2780 cells as shown by Western blotting of cytoplasmic cell lysates (Figure 4d). In addition, we detected higher nuclear expression levels for STAT3 and NF-kB in chemoresistant A2780cis compared to parental A2780 cells (Figure 4d). Thus, our microarray data from patient samples are validated in chemoresistant ovarian cancer cell line and investigations are ongoing to decipher the molecular mechanisms employed by ROR2 and GREB1 for their differential expression associated with cisplatin resistance.
Figure 4.
a. Cisplatin sensitivity testing of A2780 and A2780cis cell lines. Cells were incubated as five replicates for 3 days with increased concentrations of cisplatin as indicated and cell-viability was measured by CTG assay. EC50 was calculated using Graph Prism software and shown as approximate values.
b. ROR2 mRNA expression in platinum sensitive and resistant cell lines A2780/A2780cis (P = .0046). The graphs show mean ± SEM.
c. GREB1 mRNA expression in platinum sensitive and resistant cell lines A2780/A2780cis (P = .0012. The graphs show mean ± SEM.
d. Protein expression levels for ROR2, Wnt5a, NF-kB and STAT3 in the cytoplasmic and nuclear cell lysates of A2780 and A2780cis cell lines. Protein quantification was done using Oddyssey Licor software and normalized against the loading control.
Discussion
Due to its poor prognosis, identification of new therapeutic approaches is highly needed in ovarian cancer. Although approximately 50% of HGSC tumors with HR defects may benefit from PARP inhibitors, beyond this, identification of other commonly deregulated pathways could provide opportunities for better therapeutic interventions. As such, our goal was to examine gene and protein expression in a HGSC tumor sample cohort defined by response to platinum therapy and the level of PARP expression. Patients were carefully stratified based on their PARP levels and platinum responsiveness as previously indicated [20] and monitored for relatively long time, up to 50 months to ensure a comprehensive analysis of their disease progression.
Two functionally unrelated genes, GREB1 and ROR2 were identified in our analysis as significantly differentially expressed between high PARP/platinum-sensitive and low PARP/platinum-resistant groups.
While initially sequenced from brain tissue, human GREB1 is highly expressed in normal and neoplastic ovarian tissue and in several other hormone-responsive tissues such as breast, uterine and prostate [26], [27], [28], [29]. GREB1, along with CCND1 and MYC, are common transcription targets for E2 (17β-estradiol)-mediated proliferative responses, via ESR1 (estrogen receptor one) engagement [28]. Estrogen receptor positive (ESR1+,) breast cancers usually express GREB1, whereas in ovarian cancer, its expression could be detected in both, ESR1+ or estrogen receptor negative (ESR1−) tumors. ESR1-independent expression of GREB1 may indicate the existence of other signaling pathways for estrogen-promoting growth in the absence of E2 and/or ESR1, underlining differences in E2-responsiveness between breast and ovarian cancer that play an important role in response to antiestrogenic therapies [30]. GREB1 was upregulated in all EOC tumors and was suggested to have potential biomarker role in ovarian cancer [29]. GREB1 was upregulated in our microarray data in high PARP/platinum-sensitive patients, and high GREB1 expression was associated with longer PFS, suggesting a prognostic value for GREB1 in ovarian cancer.
It has been demonstrated that GREB1 knockdown inhibits proliferation of ovarian cancer cell lines and consequently, prolongs survival in an orthotopic mouse model [28]. Also, hypomethylation at specific CpG site associated with GREB1 has been associated with longer PFS in ovarian cancer in a DNA methylation study designed to investigate epigenetic modifications [31]. Interestingly, loss of GREB1 has been linked to tamoxifen resistance in breast cancer due to loss of sensitivity to endocrine agents in general, underlining its important role in endocrine resistance [32], [33]. Although endocrine therapy has overall limited efficacy in ovarian cancer patients, more research is needed to assess whether GREB1 is associated with antiestrogen sensitivity in this cancer. In view of this previous data, our results are in accordance with these findings reaffirming the understanding that GREB1 is highly expressed in high PARP/platinum-sensitive patient group that has been associated with a positive outcome befitting with the BRCAness profile [10]. Our data show for the first time that GREB1 could have prognostic value in ovarian cancer and warrants further investigation, especially associated with response to hormone-based therapy.
ROR2 belongs to the ROR receptor family (ROR1 and ROR2) from the non-canonical Wnt pathway [17]. ROR1 and ROR2 form heterodimers in response to Wnt5a, which leads to RhoA/Rac1 activation and increases migration and invasion properties of cancer cells [34]. Recent studies have demonstrated that upregulation of ROR2 and its ligand Wnt5a in EOC regulates EMT and correlates with worse prognosis [35]. ROR1 and ROR2 regulate migration and invasion of ovarian cancer cells and more importantly, their expression was increased in platinum resistant A2780 ovarian cancer cell line compared to parental cells ([18]). TCGA data analysis of over 500 ovarian tumor samples identified high expression of Wnt5a and Wnt5a protein was found prevalent in ascites samples of ovarian cancer patients [36]. In our study, ROR2 gene expression was significantly upregulated in low PARP/platinum-resistant vs. high PARP/platinum-sensitive patient samples. We also found higher protein levels of ROR2, ROR1 and Wnt5a ligand in lysates of platinum-resistant tumors (Figure 3, a-b), confirming our gene expression analysis. Interestingly, higher expression levels of NF-κB and pSTAT3 (Y705) proteins were also noted in platinum-resistant tumor lysate with high ROR1 and ROR2 levels, indicating that STAT3 could be downstream mediator of ROR signaling in ovarian cancer. These results showing Wnt5a-ROR2 and STAT3/NF-kB signaling pathway in clinical ovarian tumor samples are novel findings. A direct link between ROR1 and STAT3 expression has been demonstrated previously in leukemia, showing that STAT3 promoter harbors two ROR1 binding sites [37]. Moreover, previous studies have shown that Wnt5a is involved in cancer multidrug resistance (MDR)[36]. High Wnt5a expression levels in ovarian cancer cell lines correlated with lower chemosensitivity to paclitaxel, oxaliplatin, 5-fluorouracil, epirubicin and etoposide ([38], [39]. Furthermore, upregulation of Wnt5a and ROR2 was detected in colon cancer cells resistant to butyrate, a histone deacetylase inhibitor (HDACi)[40], along with activation of AKT/PKB (protein kinase B) signaling pathway. Wnt5a orchestrates multiple signaling networks involved in chronic inflammation and carcinogenesis, and crosstalk with STAT3 and NF-κB pathways have been previously documented [41]. Thus, upregulation of Wnt5a, ROR1 and ROR2 signaling pathways could be common mechanisms in ovarian cancer chemoresistance and could serve as putative biomarkers. ROR1 targeted therapies have shown promising results in preclinical and clinical models, with anti-ROR1 monoclonal antibody cirmtuzumab being efficient not only in leukemia, but also in ovarian cancer [42], indicative of high therapeutic potential for targeting these receptors.
Our mRNA expression data from patient samples were validated using a cisplatin resistant cell line model A2780cis and we found that upregulation of ROR2 at the mRNA and protein levels is associated with cisplatin resistance. Moreover, GREB1 mRNA level was downregulated in A2780cis compared to A2780 parental cells. Furthermore, we observed higher expression levels for Wnt5a, STAT3 and NF-kB proteins in chemoresistant A2780cis cells compared to parental cells, in support of our results from patient samples (Figure 3).
In conclusion, we found the expression of ROR2 and GREB1 to be associated with treatment response in HGSC. The association between Wnt5a/ROR2 expression and development of platinum resistance reported herein suggests that the Wnt5a/ROR2 pathway is potentially actionable for possible modulation of chemoresistance. Because silencing ROR1 and ROR2 restores the chemosensitivity of carboplatin-resistant ovarian cancer cells [18], a combination of ROR antagonists and chemotherapeutic agents may offer a promising treatment option. Also, our findings regarding GREB1 expression in highPARP/platinum-sensitive patients should reinforce the interest in this gene for future investigations.
Acknowledgments
Acknowledgements
We sincerely thank Mrs. Sari Toivola for her excellent assistance in laboratory.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Funding: This work was supported by Finnish Cancer Society (S02 Syöpäsäätiön käyttörahasto), Pirkanmaan Cancer Society (no specific grant number available), Academy of Finland (grant nr. 284663/275525), Ida Montini Foundation (no specific grant number available), the Doctoral Programme in Biomedicine and Biotechnology in University of Tampere (no specific grant number available) and Paulo Foundation, the Competitive Research Funding of Pirkanmaa Hospital District (no specific grant number available).
References
- 1.Nowak M, Głowacka E, Lewkowicz P, Banasik M, Szyłło K, Zimna K, Bednarska K, Klink M. Sub-optimal primary surgery leads to unfavorable immunological changes in ovarian cancer patients. Immunobiology. 2018;223:1–7. doi: 10.1016/j.imbio.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Heintz APM, Odicino F, Maisonneuve P, Quinn MA, Benedet JL, Creasman WT, Ngan HYS, Pecorelli S, Beller U. Carcinoma of the ovary. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95(Suppl 1):S161–S192. doi: 10.1016/S0020-7292(06)60033-7. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda A, Katanoda K. Five-year Relative Survival Rate of Ovarian Cancer in the USA, Europe and Japan. Jpn J Clin Oncol. 2014;44:196. doi: 10.1093/jjco/hyu007. [DOI] [PubMed] [Google Scholar]
- 4.Levanon K, Crum C, Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:5284–5293. doi: 10.1200/JCO.2008.18.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bast RC. Molecular approaches to personalizing management of ovarian cancer. Ann Oncol. 2011;22(Suppl 8):viii5–viii15. doi: 10.1093/annonc/mdr516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davar D, Beumer JH, Hamieh L, Tawbi H. Role of PARP inhibitors in cancer biology and therapy. Curr Med Chem. 2012;19:3907–3921. doi: 10.2174/092986712802002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang B. Development of synthetic lethality anticancer therapeutics. J Med Chem. 2014;57:7859–7873. doi: 10.1021/jm500415t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bast RC, Mills GB. Personalizing therapy for ovarian cancer: BRCAness and beyond. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:3545–3548. doi: 10.1200/JCO.2010.28.5791. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA, Cannistra SA. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2015;34:3617–3626. doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Oncogenes associated with drug resistance in ovarian cancer. J Cancer Res Clin Oncol. 2015;141:381–395. doi: 10.1007/s00432-014-1765-5. [DOI] [PubMed] [Google Scholar]
- 13.Yin F, Liu X, Li D, Wang Q, Zhang W, Li L. Tumor suppressor genes associated with drug resistance in ovarian cancer (Review) Oncol Rep. 2013;30:3–10. doi: 10.3892/or.2013.2446. [DOI] [PubMed] [Google Scholar]
- 14.Miow QH, Tan TZ, Ye J, Lau JA, Yokomizo T, Thiery J-P, Mori S. Epithelial-mesenchymal status renders differential responses to cisplatin in ovarian cancer. Oncogene. 2015;34:1899–1907. doi: 10.1038/onc.2014.136. [DOI] [PubMed] [Google Scholar]
- 15.Baarsma HA, Königshoff M, Gosens R. The WNT signaling pathway from ligand secretion to gene transcription: molecular mechanisms and pharmacological targets. Pharmacol Ther. 2013;138:66–83. doi: 10.1016/j.pharmthera.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Ford CE, Henry C, Llamosas E, Djordjevic A, Hacker N. Wnt signalling in gynaecological cancers: A future target for personalised medicine? Gynecol Oncol. 2016;140:345–351. doi: 10.1016/j.ygyno.2015.09.085. [DOI] [PubMed] [Google Scholar]
- 17.Endo M, Nishita M, Fujii M, Minami Y. Insight into the role of Wnt5a-induced signaling in normal and cancer cells. Int Rev Cell Mol Biol. 2015;314:117–148. doi: 10.1016/bs.ircmb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Henry CE, Llamosas E, Djordjevic A, Hacker NF, Ford CE. Migration and invasion is inhibited by silencing ROR1 and ROR2 in chemoresistant ovarian cancer. Oncogene. 2016;5:e226. doi: 10.1038/oncsis.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Cancer Policy Forum, Board on Health Care Services, Health and Medicine Division, National Academies of Sciences, Engineering, and Medicine . National Academies Press (US); Washington (DC): 2017. The Drug Development Paradigm in Oncology: Proceedings of a Workshop, The National Academies Collection: Reports funded by National Institutes of Health. [PubMed] [Google Scholar]
- 20.Veskimäe K, Staff S, Grönholm A, Pesu M, Laaksonen M, Nykter M, Isola J, Mäenpää J. Assessment of PARP protein expression in epithelial ovarian cancer by ELISA pharmacodynamic assay and immunohistochemistry. Tumour Biol. 2016;37:11991–11999. doi: 10.1007/s13277-016-5062-6. [DOI] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Mardia KV, Li Q, Hainsworth TJ. On the Penrose hypothesis on fingerprint patterns. IMA J Math Appl Med Biol. 1992;9:289–294. doi: 10.1093/imammb/9.4.289. [DOI] [PubMed] [Google Scholar]
- 25.Henry C, Llamosas E, Knipprath-Meszaros A, Schoetzau A, Obermann E, Fuenfschilling M, Caduff R, Fink D, Hacker N, Ward R. Targeting the ROR1 and ROR2 receptors in epithelial ovarian cancer inhibits cell migration and invasion. Oncotarget. 2015;6:40310–40326. doi: 10.18632/oncotarget.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hnatyszyn HJ, Liu M, Hilger A, Herbert L, Gomez-Fernandez CR, Jorda M, Thomas D, Rae JM, El-Ashry D, Lippman ME. Correlation of GREB1 mRNA with protein expression in breast cancer: validation of a novel GREB1 monoclonal antibody. Breast Cancer Res Treat. 2010;122:371–380. doi: 10.1007/s10549-009-0584-x. [DOI] [PubMed] [Google Scholar]
- 27.Fung JN, Holdsworth-Carson SJ, Sapkota Y, Zhao ZZ, Jones L, Girling JE, Paiva P, Healey M, Nyholt DR, Rogers PAW. Functional evaluation of genetic variants associated with endometriosis near GREB1. Hum Reprod. 2015;30:1263–1275. doi: 10.1093/humrep/dev051. [DOI] [PubMed] [Google Scholar]
- 28.Pellegrini C, Gori I, Achtari C, Hornung D, Chardonnens E, Wunder D, Fiche M, Canny GO. The expression of estrogen receptors as well as GREB1, c-MYC, and cyclin D1, estrogen-regulated genes implicated in proliferation, is increased in peritoneal endometriosis. Fertil Steril. 2012;98:1200–1208. doi: 10.1016/j.fertnstert.2012.06.056. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkinson KM. Department of Cellular and Molecular Medicine University of Ottawa; Ottawa, Ontario, Canada: 2016. The role of steroid hormones, GREB1, and reproductive status in ovarian cancer progression. [Google Scholar]
- 30.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauerschlag DO, Ammerpohl O, Bräutigam K, Schem C, Lin Q, Weigel MT, Hilpert F, Arnold N, Maass N, Meinhold-Heerlein I. Progression-free survival in ovarian cancer is reflected in epigenetic DNA methylation profiles. Oncology. 2011;80:12–20. doi: 10.1159/000327746. [DOI] [PubMed] [Google Scholar]
- 32.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JLL. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Zhang Z, Cenciarini ME, Proietti CJ, Amasino M, Hong T, Yang M, Liao Y, Chiang H-C, Kaklamani VG. Tamoxifen Resistance in Breast Cancer Is Regulated by the EZH2-ERα-GREB1 Transcriptional Axis. Cancer Res. 2018;78:671–684. doi: 10.1158/0008-5472.CAN-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, Chen L, Cui B, Widhopf GF, Shen Z, Wu R, Zhang L, Zhang S, Briggs SP, Kipps TJ. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Invest. 2016;126:585–598. doi: 10.1172/JCI83535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ford CE, Punnia-Moorthy G, Henry CE, Llamosas E, Nixdorf S, Olivier J, Caduff R, Ward RL, Heinzelmann-Schwarz V. The non-canonical Wnt ligand, Wnt5a, is upregulated and associated with epithelial to mesenchymal transition in epithelial ovarian cancer. Gynecol Oncol. 2014;134:338–345. doi: 10.1016/j.ygyno.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Asem M, Buechler S, Wates R, Miller D, Stack M. Wnt5a Signaling in Cancer. Cancer. 2016;8:79. doi: 10.3390/cancers8090079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li P, Harris D, Liu Z, Liu J, Keating M, Estrov Z. Stat3 activates the receptor tyrosine kinase like orphan receptor-1 gene in chronic lymphocytic leukemia cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng C, Zhang X, Yu H, Wu D, Zheng J. Wnt5a as a predictor in poor clinical outcome of patients and a mediator in chemoresistance of ovarian cancer. Int J Gynecol Cancer. 2011;21:280–288. doi: 10.1097/IGC.0b013e31820aaadb. [DOI] [PubMed] [Google Scholar]
- 39.Varma RR, Hector SM, Clark K, Greco WR, Hawthorn L, Pendyala L. Gene expression profiling of a clonal isolate of oxaliplatin-resistant ovarian carcinoma cell line A2780/C10. Oncol Rep. 2005;14:925–932. [PubMed] [Google Scholar]
- 40.Bordonaro M, Tewari S, Cicco CE, Atamna W, Lazarova DL. A switch from canonical to noncanonical Wnt signaling mediates drug resistance in colon cancer cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh Transcriptional mechanisms of WNT5A based on NF-κB, Hedgehog, TGFβ, and Notch signaling cascades. Int J Mol Med. 2009;23 doi: 10.3892/ijmm_00000190. [DOI] [PubMed] [Google Scholar]
- 42.Choi MY, Widhopf GF, Wu CCN, Cui B, Lao F, Sadarangani A, Cavagnaro J, Prussak C, Carson DA, Jamieson C. Pre-clinical Specificity and Safety of UC-961, a First-In-Class Monoclonal Antibody Targeting ROR1. Clin Lymphoma Myeloma Leuk. 2015;15(Suppl):S167–S169. doi: 10.1016/j.clml.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]