Figure 1.
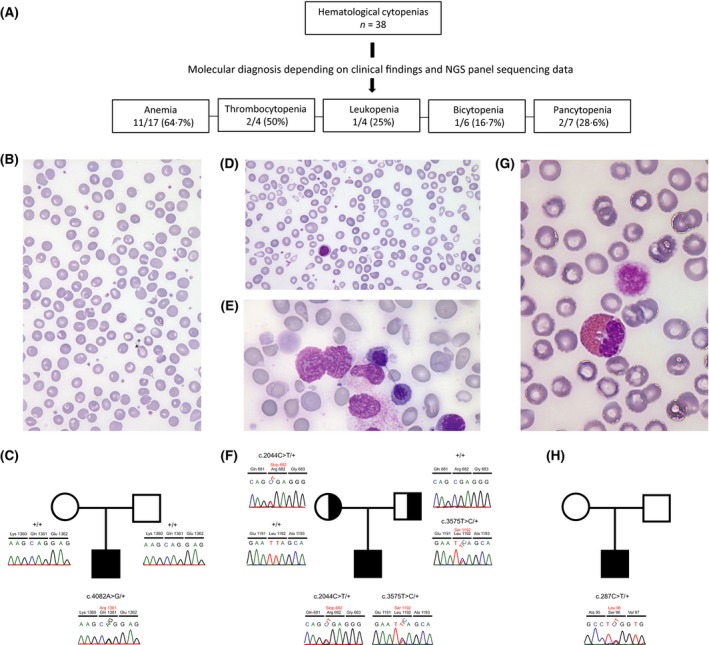
Targeted NGS‐based panel screen for efficient identification of genetic causes in inherited haematological diseases. (A) A total of 38 patients with molecularly undefined cytopenias were included in the study, and a disease‐causing mutation was identified in 17 (44·7%). (B) May‐Grünwald‐Giemsa stained peripheral blood smear showing characteristic dessicytes and a few target cells in Patient 8 with hereditary xerocytosis caused by (C) a novel mutation in PIEZO1 (NM_001142864.2:c.4082A>G p.Q1361R). In Patient 9, who exhibited characteristic light microscopy findings in (D) peripheral blood, i.e., gross anisocytosis, poikilocytosis and (E) bone marrow, such as erythroid hyperplasia and chromatin bridges between nuclei of two separate erythroblasts, we identified (F) novel compound heterozygous variants (NM_138477.2:c.2044C>T p.R682X and NM_138477.2:c.3575T>C p.L1192S) in codanin 1 (CDAN1) causing congenital dyserythropoietic anaemia type 1. In an infant (Patient 13) with thrombocytopenia and giant platelets (G) [giant platelet (right) and eosinophilic granulocyte (left)], we identified a (H) known causative mutation in MHY9 (NM_002473.4:c.287C>T p.S96L) causing MYH9‐related disease. NGS, next generation sequencing.
