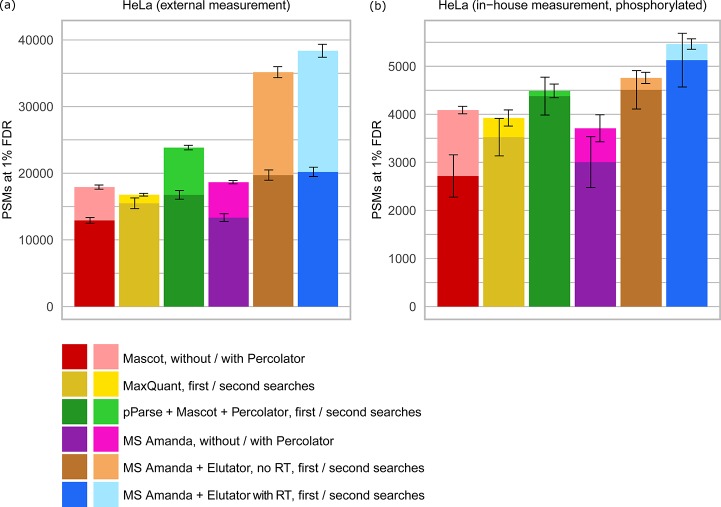
Figure 4.
Comparison of MS Amanda and Elutator with other scoring methods and validation tools. Comparison was performed using (a) an external HeLa data set obtained from Michalski et al.32 (data set H) and (b) an in-house data set of human HeLa after TiO2 enrichment of phosphorylated peptides (data set G). The FDR threshold of 1% was calculated at PSM level for consistency between different search tools, which typically operate at PSM level. In cases where several high confident matches were reported for the same spectrum, the match with best q-value was selected such that the number of PSMs corresponds to the number of confidently identified spectra. Elutator includes features derived from a peptide elution retention time prediction model. Model training was performed on in–house data sets, the same model was applied to in-house and external data sets.