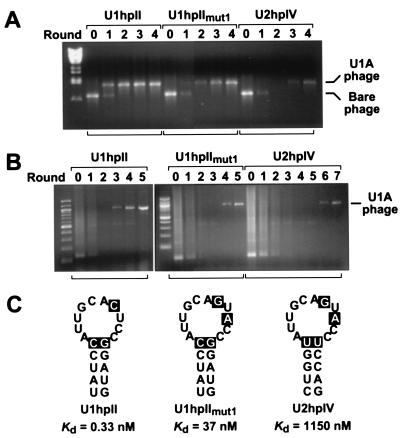
Figure 3.
Selection of U1A-displaying phage from phage pools of various complexities. (A) Selection of recombinant U1A-displaying phage after dilution with a 106-fold excess of bare phage. The phage mixture (2 × 109 pfu) was incubated with each of three bait RNAs (100 nM): U1hpII, a U1hpII variant (U1hpIImut1), or U2hpIV. Multiple rounds of in vitro selection were performed as outlined in Fig. 1. The relative abundance of U1A-displaying phage in the original phage mixture (denoted round 0) and after each round of selection was monitored by PCR analysis with primers flanking the site of cDNA insertion. Signals corresponding to the U1A-displaying phage and the bare phage are indicated. (B) Isolation of U1A-displaying phage from a human lung cDNA library comprising 1.2 × 107 independent clones. In vitro selection from the phage library (2.5 × 109 pfu) was performed as in A, using the same three bait RNAs (100 nM) but a different reverse primer for PCR analysis. The cDNA insert of the phage clone isolated in this manner was shown by sequence analysis to encode full-length U1A. (C) Schematic representation of the three RNA stem loop structures used in these experiments. The previously determined affinity of each for U1A (Kd) is indicated (23). The nucleotide changes in U1hpIImut1 and U2hpIV that are responsible for the diminished affinity of U1A for these stem loops are highlighted. In addition to the stem loop structures shown, each RNA contained four or six additional base pairs at the bottom of the stem, a short 5′ leader, and a 3′ tail that had been annealed to a complementary 5′-biotinylated DNA oligonucleotide before selection.