Abstract
Aflatoxins and ochratoxins are among the most important mycotoxins of all and producers of both types of mycotoxins are present in Aspergillus section Flavi, albeit never in the same species. Some of the most efficient producers of aflatoxins and ochratoxins have not been described yet. Using a polyphasic approach combining phenotype, physiology, sequence and extrolite data, we describe here eight new species in section Flavi. Phylogenetically, section Flavi is split in eight clades and the section currently contains 33 species. Two species only produce aflatoxin B1 and B2 (A. pseudotamarii and A. togoensis), and 14 species are able to produce aflatoxin B1, B2, G1 and G2: three newly described species A. aflatoxiformans, A. austwickii and A. cerealis in addition to A. arachidicola, A. minisclerotigenes, A. mottae, A. luteovirescens (formerly A. bombycis), A. nomius, A. novoparasiticus, A. parasiticus, A. pseudocaelatus, A. pseudonomius, A. sergii and A. transmontanensis. It is generally accepted that A. flavus is unable to produce type G aflatoxins, but here we report on Korean strains that also produce aflatoxin G1 and G2. One strain of A. bertholletius can produce the immediate aflatoxin precursor 3-O-methylsterigmatocystin, and one strain of Aspergillus sojae and two strains of Aspergillus alliaceus produced versicolorins. Strains of the domesticated forms of A. flavus and A. parasiticus, A. oryzae and A. sojae, respectively, lost their ability to produce aflatoxins, and from the remaining phylogenetically closely related species (belonging to the A. flavus-, A. tamarii-, A. bertholletius- and A. nomius-clades), only A. caelatus, A. subflavus and A. tamarii are unable to produce aflatoxins. With exception of A. togoensis in the A. coremiiformis-clade, all species in the phylogenetically more distant clades (A. alliaceus-, A. coremiiformis-, A. leporis- and A. avenaceus-clade) are unable to produce aflatoxins. Three out of the four species in the A. alliaceus-clade can produce the mycotoxin ochratoxin A: A. alliaceus s. str. and two new species described here as A. neoalliaceus and A. vandermerwei. Eight species produced the mycotoxin tenuazonic acid: A. bertholletius, A. caelatus, A. luteovirescens, A. nomius, A. pseudocaelatus, A. pseudonomius, A. pseudotamarii and A. tamarii while the related mycotoxin cyclopiazonic acid was produced by 13 species: A. aflatoxiformans, A. austwickii, A. bertholletius, A. cerealis, A. flavus, A. minisclerotigenes, A. mottae, A. oryzae, A. pipericola, A. pseudocaelatus, A. pseudotamarii, A. sergii and A. tamarii. Furthermore, A. hancockii produced speradine A, a compound related to cyclopiazonic acid. Selected A. aflatoxiformans, A. austwickii, A. cerealis, A. flavus, A. minisclerotigenes, A. pipericola and A. sergii strains produced small sclerotia containing the mycotoxin aflatrem. Kojic acid has been found in all species in section Flavi, except A. avenaceus and A. coremiiformis. Only six species in the section did not produce any known mycotoxins: A. aspearensis, A. coremiiformis, A. lanosus, A. leporis, A. sojae and A. subflavus. An overview of other small molecule extrolites produced in Aspergillus section Flavi is given.
Key words: Aspergillus, Section Flavi, Aflatoxins, Cyclopiazonic acid, Tenuazonic acid
Taxonomic novelties: Aspergillus aflatoxiformans Frisvad, Ezekiel, Samson & Houbraken, Aspergillus aspearensis Houbraken, Frisvad, Arzanlou & Samson, Aspergillus austwickii Frisvad, Ezekiel, Samson & Houbraken, Aspergillus cerealis Houbraken, Frisvad, Ezekiel & Samson, Aspergillus neoalliaceus A. Nováková, Hubka, Samson, Frisvad & Houbraken, Aspergillus pipericola Frisvad, Samson & Houbraken, Aspergillus subflavus Hubka, A. Nováková, Samson, Frisvad & Houbraken, A. vandermerwei Frisvad, Hubka, Samson & Houbraken
Introduction
Aspergillus subgenus Circumdati section Flavi contains some of the most important species in the genus, which are of significance in biotechnology, foods and health (Varga et al. 2011). Aspergillus flavus is reported, after A. fumigatus (section Fumigati), as the second leading cause of invasive aspergillosis and it is the most common cause of superficial infection (Hedayati et al. 2007). Aspergillus oryzae and A. sojae appear to be the domesticated forms of the aflatoxigenic species A. flavus and A. parasiticus, respectively, and are used extensively in the food and biotechnology industries (Houbraken et al. 2014). A large number of species in Aspergillus section Flavi are common in crops, and some of them produce several mycotoxins, such as aflatoxins, 3-nitropropionic acid, tenuazonic acid and cyclopiazonic acid (Varga et al. 2011). Despite many publications in various research fields, the taxonomy of the aflatoxigenic species in Aspergillus section Flavi is still not fully elucidated, and several new species (some with aflatoxigenic potential) have been described since 2011, such as A. novoparasiticus (Gonçalves et al., 2012a, Gonçalves et al., 2012b), A. mottae, A. transmontanensis, A. sergii (Soares et al. 2012), A. bertholletius (Taniwaki et al. 2012), A. hancockii (Pitt et al. 2017) and A. korhogoensis (Carvajal-Campos et al. 2017). Additionally, there have also been some disagreements on the proper species names of strains formerly identified as A. flavus with large or small sclerotia (Probst et al., 2012, Probst et al., 2014).
Initially, A. flavus was reported to produce aflatoxin of the B and G type (Nesbitt et al., 1962, Codner et al., 1963). Later it was recognised that strains of A. flavus can only produce aflatoxin B1 and B2 (Varga et al., 2009, Amaike and Keller, 2011) and that the strains producing aflatoxin B and G were A. parasiticus, exemplified by strain NRRL 2999, which was initially identified as A. flavus (Christensen et al. 1973) and three years later re-identified as A. parasiticus (Buchanan & Ayres 1976). Although it was considered that A. flavus only produces B type aflatoxins, some reports indicate that A. flavus strains can also produce the G type aflatoxins (Camiletti et al., 2017, Barayani et al., 2015, Wicklow and Shotwell, 1983, Okoth et al., 2018, Saldan et al., 2018). This contradictory data needs further investigation and it is important to determine whether A. flavus sensu stricto can produce aflatoxins of the G type or not. Most species in Aspergillus section Flavi produce both types of aflatoxins, while species outside section Flavi can only accumulate sterigmatocystin and aflatoxins of the B type (Geiser et al., 2007, Varga et al., 2009, Rank et al., 2011).
Raper & Fennell (1965) stated that A. flavus strains produced globose to subglobose sclerotia that are normally 400–700 μm in size, rarely exceeding 1 mm, but that some strains produced sclerotia that were uniformly and consistently smaller. They also mentioned strains that produced vertically elongate sclerotia, and such strains were later shown to be A. nomius or A. pseudonomius (Kurtzman et al. 1997, Varga et al., 2011, Massi et al., 2014). Also Hesseltine et al. (1970) reported A. flavus isolates with small sclerotia while most isolates had large sclerotia. They listed NRRL 3251 as one of the rare examples of a strain with small sclerotia that produced aflatoxin B1 and B2 only, and stated that this could represent a new species. Another strain similar to NRRL 3251 that also produce small-sized sclerotia is the genome sequenced strain ATCC MYA384 (= AF70) (Moore et al. 2015). These A. flavus strains with small sclerotia that produce B type aflatoxins (A. flavus SB) are more common in USA than in Africa (Probst et al. 2014). Later Saito & Tsuruta (1993) found many strains with small sclerotia from agricultural soil in Thailand. They described their strains and NRRL 3251 as A. flavus var. parvisclerotigenus. In 2005, Frisvad et al. (2005) raised A. flavus var. parvisclerotigenus to species level and neotypified the species with a strain isolated from a peanut in Nigeria producing aflatoxin B1, B2, G1 and G2 (CBS 121.62 = IMI 093070 = NRRL A-11612). This neotypification is questionable as the original type only produced B type aflatoxins. Other strains producing small sclerotia, often referred to as A. flavus group SBG (= “A. flavus strains producing small sized sclerotia and aflatoxin B and G”) represent multiple species. One of the “A. flavus group SBG” taxa was described as A. minisclerotigenes (from Argentina originally) (Pildain et al. 2008) and is also found in Central, East and Southern Africa and Australia (Probst et al. 2014), while A. parvisclerotigenus sensu Frisvad et al. (2005) has been found in West Africa: Benin, Burkina Faso, Nigeria, Senegal and Sierra Leone (Probst et al. 2014). Another important group of strains is identified as A. flavus SB and these strains are regarded as the agent causing lethal levels of aflatoxins in Kenyan maize. It remains questionable whether these are truly A. flavus or that these strains represent a species that has not yet been named (Cotty and Cardwell, 1999, Cardwell and Cotty, 2002, Donner et al., 2009, Okoth et al., 2012, Okoth et al., 2018, Probst et al., 2007, Probst et al., 2010, Probst et al., 2012, Probst et al., 2014). However, a later study shows A. flavus sensu stricto and A. minisclerotigenes are the predominant species in Kenyan maize (Okoth et al. 2018).
The genomes of A. oryzae RIB 40 (Machida et al., 2005, Galagan et al., 2005, Inglis et al., 2013, Umemura et al., 2013a, Umemura et al., 2013b), and other strains of A. oryzae (Zhao et al., 2012, Zhao et al., 2013, Zhao et al., 2014), A. flavus NRRL 3357 (= ATCC 200026) (Payne et al., 2006, Fedorova et al., 2008, Nierman et al., 2015), ATCC MYA384 (= AF70) (Moore et al. 2015) and other strains (Faustinelli et al. 2016), A. parasiticus ATCC 56775 (= NRRL 5862 = SU-1) (Linz et al. 2014), A. sojae NBRC 4239 (Sato et al. 2011), A. bombycis NRRL 26010 (Moore et al. 2016), A. nomius NRRL 13137 (= NBRC 33223) (Horn et al., 2009c, Moore et al., 2015), A. hancockii FRR 3425 (Pitt et al. 2017) and A. arachidicola (Moore et al. 2018) have been published. Gene clusters for several secondary metabolites, and the regulation of these gene clusters in A. flavus are known, including those for aflatoxins, aflatrem, aflavarins, aflavinines, asparasones, cyclopiazonic acid, kojic acid, leporins and penicillin (Chang et al., 2009, Geogianna et al., 2010, Marui et al., 2010, Terebayashi et al., 2010, Chang and Ehrlich, 2011, Marui et al., 2011, Amare and Keller, 2014, Ehrlich and Mack, 2014, Tang et al., 2015, Cary et al., 2015a, Cary et al., 2015b, Cary et al., 2017, Gilbert et al., 2016, Ammar et al., 2017, Chang et al., 2017, Ibarra et al., 2018). Genome sequencing of more strains in section Flavi will help elucidating how the gene clusters for aflatoxins and ochratoxins evolved. Sexual reproduction appears to be important for the variation between isolates of A. flavus, so acquisition of new alleles and mitochondrial inheritance are factors that should be taken into consideration (Horn et al. 2016).
For food safety purposes, correct species identification is of high importance (Kim et al., 2014, Samson et al., 2006, Probst et al., 2007, Probst et al., 2010, Probst et al., 2012, Probst et al., 2014, Varga et al., 2011), as different species may have different mycotoxin profiles and physiology. For example, A. flavus strains used to prevent aflatoxin production in crops, themselves unable to produce aflatoxins, may produce other potentially toxic secondary metabolites (Ehrlich, 2014). Detection of these species in foods using sophisticated analytical techniques requires an accurate and reliable taxonomic system (Frisvad et al., 2007, Godet and Munaut, 2010, Luo et al., 2014a, Luo et al., 2014b, Faustinelli et al., 2017, Kaya-Celiker et al., 2015). Occasionally, strains producing important mycotoxins are apparently misidentified. An example of a dubious link between fungal species and mycotoxins is the production of the A. fumigatus metabolites fumigaclavine A (Jahardhanan et al. 1984) and fumitremorgins (Ma et al. 2016) by an A. tamarii strain. There is evidence that aflatoxigenic species can hybridize (Olarte et al., 2012, Olarte et al., 2015), so it should be examined whether some of the species producing aflatoxins may be hybrids. Furthermore, cells of A. flavus are multinucleate (Runa et al. 2015), and it is unknown whether such nuclei contain the same genetic material.
In this manuscript we present an update on the taxonomy of section Flavi and describe eight new species using a polyphasic approach combining physiology, morphology, sequence and extrolite data. A list of accepted species (and their synonyms) belonging to section Flavi is presented. The ability of the new species to produce aflatoxin and ochratoxin A is studied and an overview on the mycotoxin producing potential of all section Flavi species is presented.
Materials and methods
Isolation of microfungi
A part of the strains used in the study was recently isolated during various surveys in different countries (Czech Republic, Nigeria, Iran). Soil and drillosphere (soil in immediate proximity of earthworm burrows) samples and samples from Allolobophora hrabei casts and intestines were collected in 2011–2013, always in spring and autumn, in the period of earthworm activity. Soil and drillosphere samples were collected by soil corer from the top 5 cm soil layer and A. hrabei casts were collected from the soil surface. Microscopic fungi were isolated by a dilution plate method (dilution 104) and a soil washing technique (Garrett, 1981, Kreisel and Schauer, 1987) using three isolation media: dichloran rose bengal chloramphenicol agar (DRBC), Sabouraud's glucose agar (SGA) and beer wort agar (BWA). Rose bengal and chloramphenicol were added to the two latter media to suppress bacterial growth (Atlas 2010). Isolation from the A. hrabei intestine was done according to Nováková & Pižl (2003). Agar media were incubated for 7 days at 25 °C in darkness. For cultures from Nigeria, food (local rice, maize, mushroom, peanut cake and sesame) samples from various markets and agricultural soil samples from the top 2 cm of the soil were collected between October 2010 and February 2012. Cultures from food samples, except those from local rice and maize, were reported in other studies (Ezekiel et al., 2013a, Ezekiel et al., 2013b, Ezekiel et al., 2014). The isolates were previously reported as unnamed taxon SBG based on phenotype (macro- and microscopic characters on 5/2 agar (5 % V-8 juice and 2 % agar, pH 5.2)) and aflatoxin production (on neutral red desiccated coconut agar) or A. parvisclerotigenus based on ITS, β-tubulin and calmodulin gene sequences. Local rice and maize grains were milled while soil was sieved prior to isolation of moulds on modified Rose Bengal Agar (mRBA; Cotty 1994) by dilution plating (Samson et al. 1995). Cultures on mRBA and 5/2 agar were incubated for 3 and 5 days, respectively, at 31 °C in darkness. Isolated colonies were purified on 5/2 agar. For cultures from Iran, soil samples were collected at 10–15 cm depth from Aspear Island in Urmia Lake, during 2011 and 2012. Isolations were carried out using the soil dilution plate on three culture media: malt extract agar (MEA), glucose peptone yeast extract agar (GPY) and potato dextrose agar (PDA) supplemented with various NaCl concentrations (0 to 3 %) (Arzanlou et al. 2016).
Strains
The recently isolated strains (see previous paragraph) were supplemented with strains from the 1) CBS culture collection, housed at the Westerdijk Fungal Biodiversity Institute, 2) CCF, Culture Collection of Fungi, Prague, Czech Republic, 3) DTO, the working collection of the Applied and Industrial Mycology department housed at the Westerdijk Institute, 4) IBT, the culture collection of at the Department of Biotechnology and Biomedicine, Technical University of Denmark, 5) KACC, Korean Agricultural Culture Collection, Wanju, South Korea. Interesting strains and strains representing new species were deposited into the public CBS culture collection.
Morphological characterisation
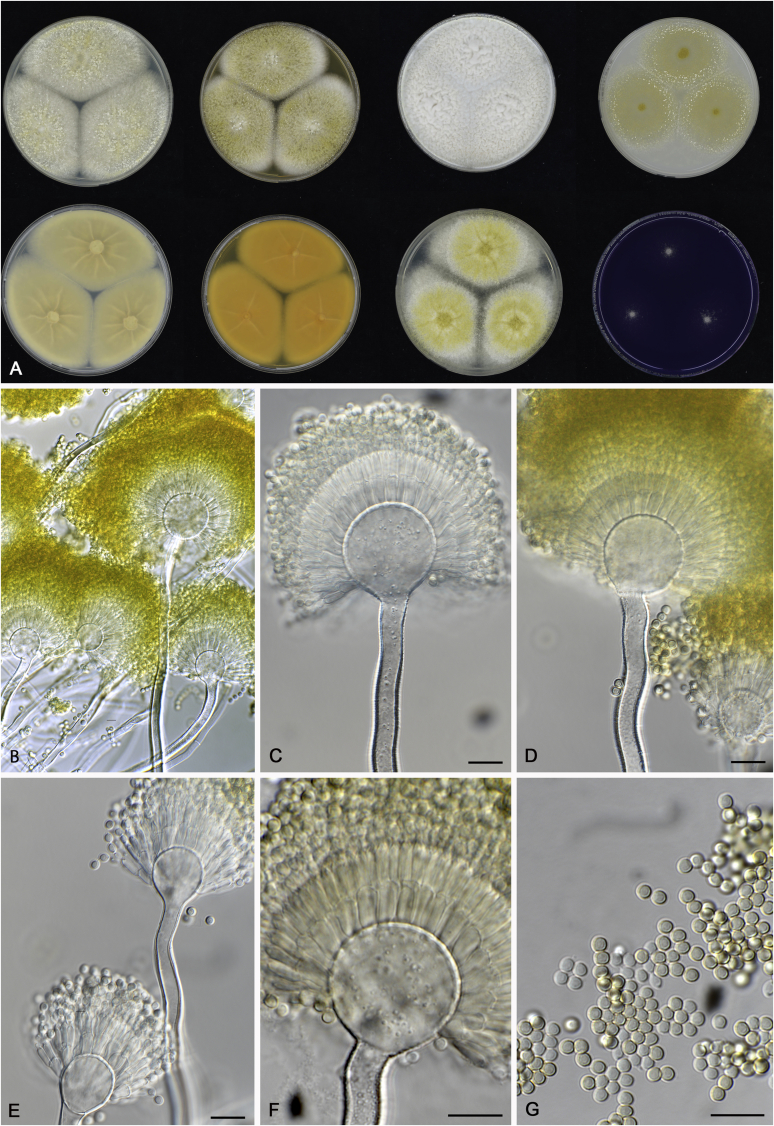
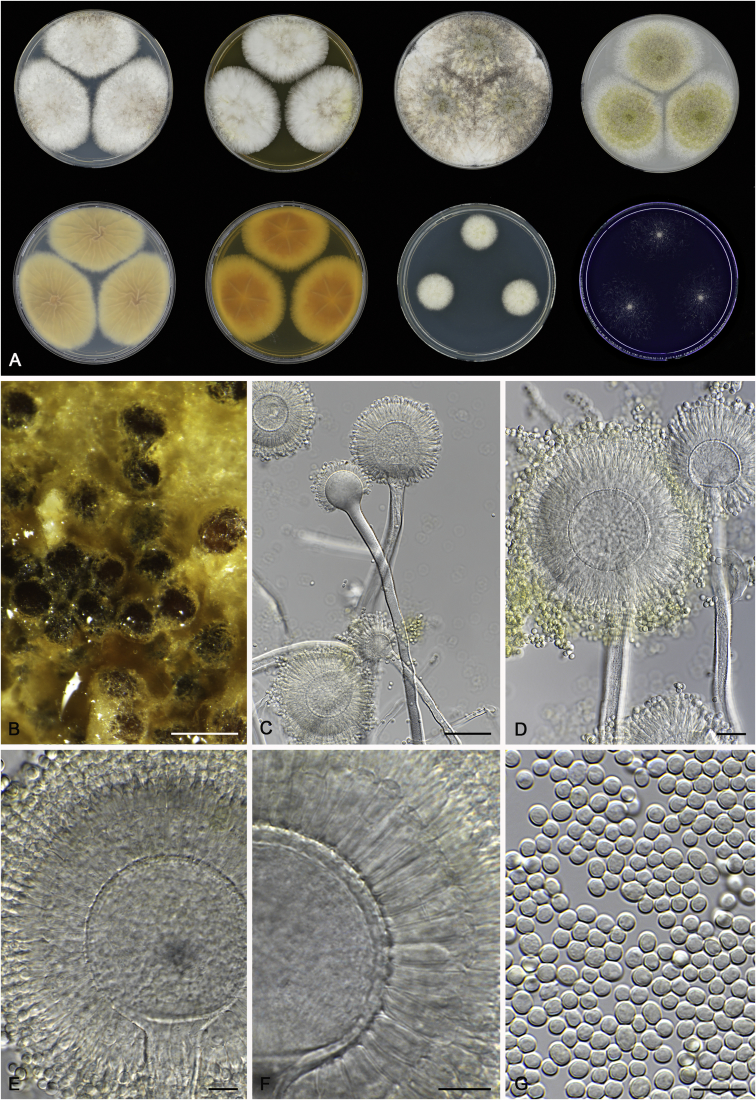
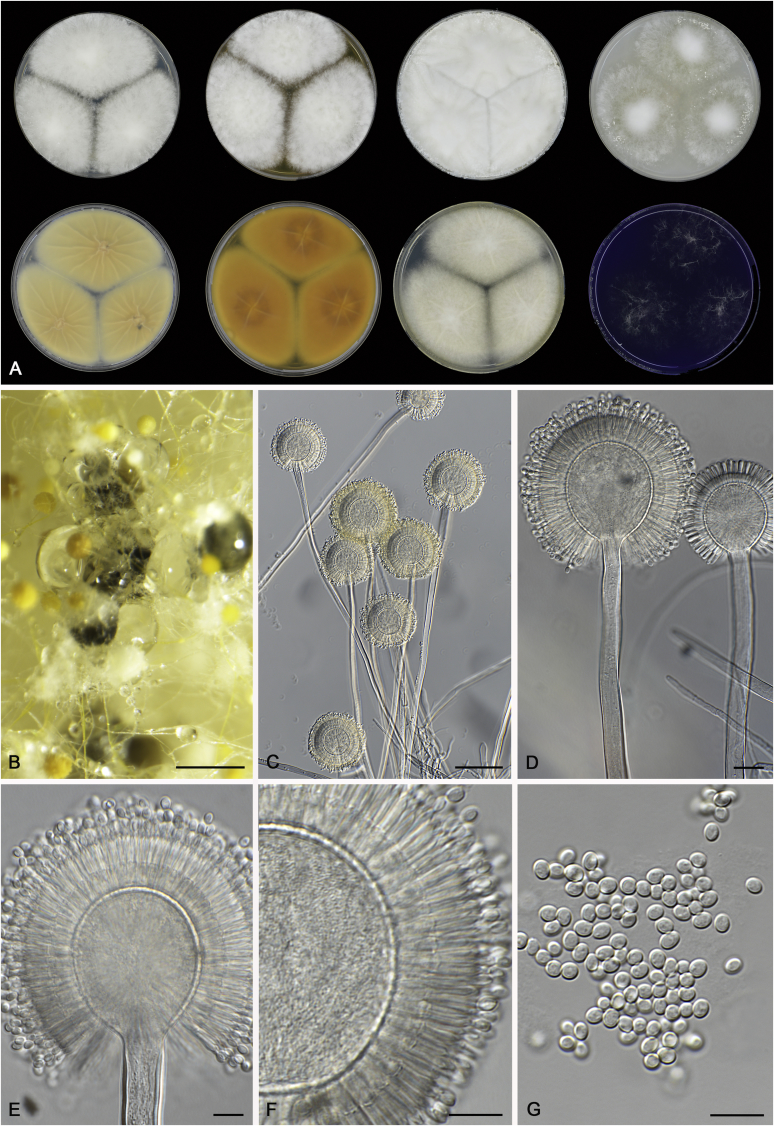
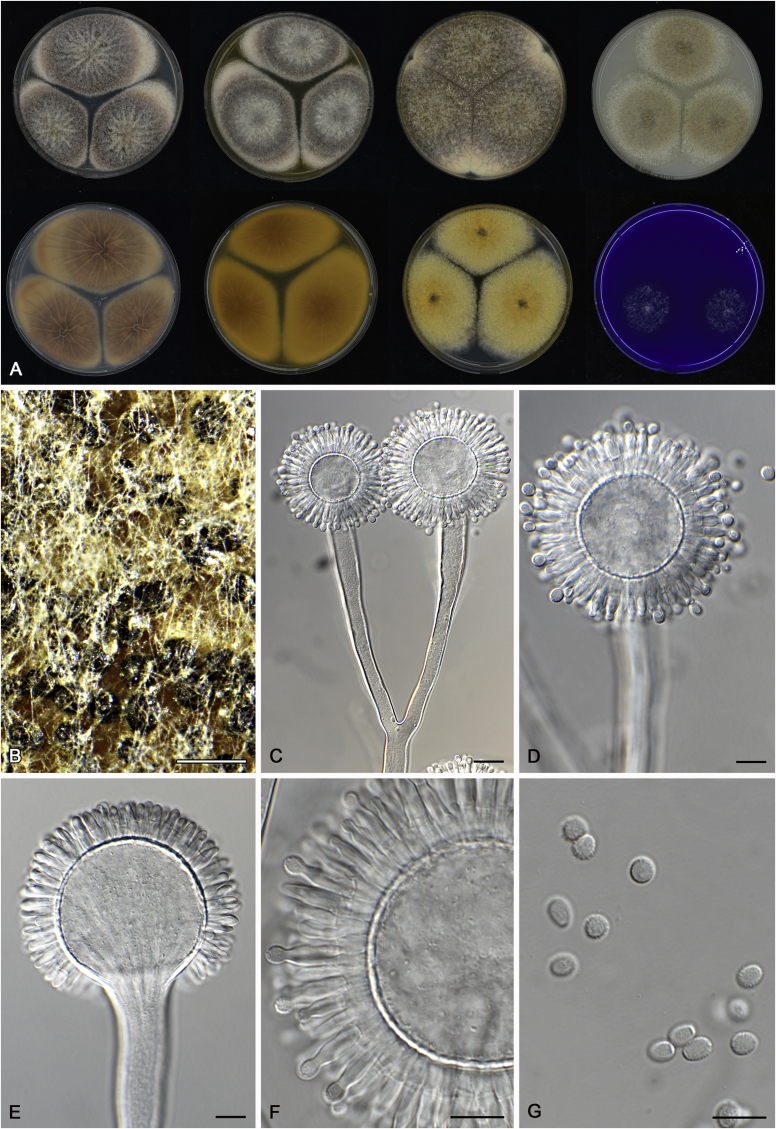
Cultures for macromorphological observations were inoculated in a three point position onto the agar media creatine sucrose agar (CREA), Czapek yeast extract agar (CYA), CYA supplemented with 5 % NaCl (CYAS), dichloran 18 % glycerol agar (DG18), malt extract agar (Oxoid) (MEA), oatmeal agar (OA) and yeast extract sucrose agar (YES). All media were prepared as described by Samson et al. (2014). Additional CYA plates were inoculated and incubated at 37 °C (CYA37°C) and 42 °C (CYA42°C). Colony texture, degree of sporulation, obverse and reverse colony colours, production of soluble pigments, exudates and sclerotia/ascomata were determined and recorded after 7 d of incubation. Colours names and codes used in descriptions refer to Rayner (1970). The production of sclerotia was observed with the naked eye. Digital images of these structures were made from CYA plates (incubate at 25 or 37 °C) and captured using a Nikon SMZ25 dissecting microscope. For micromorphological observations, mounts were made in lactic acid (60 %) from colonies on MEA and a drop of ethanol was used to wash excess conidia. The possible production of a sexual state was observed on OA, MEA and CYA plates incubated up to six weeks. Structures were studied and captured by using a Zeiss AX10 Imager A2 light microscope equipped with a Nikon DS-Ri2 camera and the software package NIS-Elements D v4.50. Photoplates were prepared in Adobe® Photoshop® CS6.
DNA extraction, amplification and sequencing
DNA was extracted from 3–7 days-old colonies with the DNA extraction kit ArchivePure DNA yeast and Gram2+ kit (5PRIME Inc., Gaithersburg, Maryland) with modifications described by Hubka et al. (2015) or the UtracleanTM Microbial DNA isolation kit (MoBio, Solana Beach U.S.A.). The ITS rDNA region was amplified using forward primers ITS1 and ITS5 (White et al. 1990) and reverse primers ITS4S (Kretzer et al. 1996 or NL4 (O’Donnell 1993), or the primer pair V9G (de Hoog & Gerrits van den Ende, 1998) and LS266 (Masclaux et al. 1995); a part of the BenA gene encoding β-tubulin using the forward primers Bt2a (Glass & Donaldson 1995) or Ben2f (Hubka & Kolařík 2012) and the reverse primer Bt2b (Glass & Donaldson 1995); partial CaM gene encoding calmodulin using forward primers CF1M or CF1L and reverse primer CF4 (Peterson 2008), or the primer pair cmd5 and cmd6 (Hong et al. 2005); partial RPB2 gene using forward primers fRPB2-5F (Liu et al. 1999), RPB2-F50-CanAre (Jurjević et al. 2015), RPB2-5F_Eur (Houbraken et al. 2012) and reverse primers RPB2-7CR_Eur (Houbraken et al. 2012) and fRPB2-7cR (Liu et al. 1999). PCR protocols were described by Hubka et al., 2014, Hubka et al., 2016 and Samson et al. (2014). Automated sequencing was performed with the same primers as used in PCR reactions.
Phylogenetic analysis
The sequence data were inspected, assembled and optimised using the software package Seqman (v. 10.0.1) from DNAStar Inc. Sequences were aligned with MAFFT v.7 (Katoh & Standley 2013) using the L-INS-i method. Maximum likelihood (ML) analysis on the combined data sets was performed using the RAxML v. 7.2.6 (randomized axalerated maximum likelihood) software (Stamatakis & Alachiotis 2010). The combined data sets were analysed as three distinct partitions (BenA, CaM and RPB2). For each individual data set, the most optimal substitution model was calculated in the MEGA7 v. 7.0.25 software (Kumar et al. 2016) utilising the Akaike Information Criterion (AIC). Maximum Likelihood analysis of the individual data sets was analysed performed using MEGA7 and the robustness of the trees was evaluated by 1 000 bootstrap replicates. A second measure for statistical support was performed using MrBayes v. 3.2.2 (Ronquist et al. 2012) and the previously obtained most optimal substitution model was used in the analyses. The Markov Chain Monte Carlo (MCMC) analysis used four chains and started from a random tree topology. Burn-in was set to 25 % and Tracer v. 1.5.0 (Rambaut & Drummond 2009) was used to confirm the convergence of chains. The phylograms obtained during the ML analysis were used for presenting the data. Phylograms were redrawn from the tree files using TREEVIEW (Page 1996) and optimized using Adobe® Illustrator® CS5.1. Bootstrap (BS) values lower than 70 % and posterior probability (pp) values lower than 0.95 were removed from the phylograms. The phylogenetic relationship of species belonging to section Flavi is studied using a combined data set of partial BenA, CaM and RPB2 gene sequences. The relationship of strains (and species) belonging to five clades (A. alliaceus-, A. flavus-, A. leporis-, A. nomius- and A. tamarii-clade) is studied in more detail. The reason of these detail analyses is that either these clades contain new species and/or we found that the taxonomy of those clades was not well re-solved. Aspergillus muricatus NRRL 35674T was used as out-group in the overview of section Flavi, Aspergillus tamarii NRRL 20818 in the A. flavus- and A. leporis-clade phylogenies and A. bertholletius CBS 143687T in the A. alliaceus-, A. nomius- and A. tamarii-clade phylogenies.
Extrolite analysis
Strains were grown for 7 d at 25 °C on YES and CYA prior to extrolite extraction. The strains of the recently described species were inoculated on Czapek yeast autolysate (CYA) agar, malt extract agar (MEA) (Blakeslee formula), MEA-Ox (Oxoid formula), yeast extract sucrose (YES) agar, oat meal (OAT) agar, potato dextrose agar (PDA) (Difco), Wickerhams antibiotic test medium (WATM) and Raulin Thom oat meal (RTO) agar (Nielsen et al., 2011a, Nielsen et al., 2011b, Frisvad, 2012), and Aspergillus flavus parasiticus agar (AFPA) (Pitt et al. 1983).
Strains listed in Table 1 were tested for production of small molecule extrolites according to the agar plug extraction method of Filtenborg et al. (1983) as modified by Smedsgaard (1997). The HPLC-DAD method was following Frisvad & Thrane (1987), as modified by Nielsen & Smedsgaard (2003) and Nielsen et al., 2011a, Nielsen et al., 2011b. After extracting the agar plugs with ethylacetate /dichloromethane / methanol (3:2:1, vol/vol/vol) containing 1 % formic acid, the solvent was evaporated and the mixture of extrolites were re-dissolved in methanol, filtered and 1 μl was injected into a Agilent high performance liquid chromatograph with a diode array detector. Samples made after 2015 were extracted with ethylacetate / isopropanol (3:1, vol/vol), with 1 % formic acid. Selected strains were analyzed by Ultra high performance liquid chromatography-diode array detection-high resolution quadrupole time of flight mass spectrometry (UHPLC-DAD-HRQTOFMS) according to the method of Kildgaard et al. (2014) and Klitgaard et al. (2014) using an Agilent Infinity 1290 HPLC system (Agilent Technologies, Santa Clara, CA, USA) as described in detail by Kildgaard et al. (2014).
Table 1.
Isolates examined belonging to Aspergillus section Flavi.
| Species | Isolate number | Provenance | GenBank accession no. |
|||
|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | |||
| Aspergillus aflatoxiformans | CBS 143679 = DTO 228-G2T = IBT 32085 | Agricultural soil, Minna, Niger State, Nigeria, ex type of Aspergillus aflatoxiformans | MG662388 | MG517706 | MG518076 | MG517897 |
| CBS 121.62 = IMI 093070 = NRRL A-11612 = IBT 3651 = IBT 3850 = DTO 010-H7 = DTO 223-C2 = DTO 228-H6 | Arachis hypogea, Nigeria, PKC Austwick, 1962 (former suggested neotype of Aspergillus parvisclerotigenus) | EF409240 | MG517719 | MG518089 | MG517910 | |
| CBS 133264 = DTO 215-F3 | Edible mushroom, Lagos State, Nigeria | – | JX627690 | JX627694 | MG517871 | |
| CBS 133265 = DTO 215-F4 | Edible mushroom, Lagos State, Nigeria | – | JX627691 | JX627695 | MG517872 | |
| CBS 133923 = DTO 215-F1 | Peanut cake, Niger State, Minna, Nigeria | – | MG517680 | MG518051 | MG517869 | |
| CBS 133924 = DTO 215-F2 | Peanut cake, Niger State, Minna, Nigeria | – | MG517681 | MG518052 | MG517870 | |
| CBS 133925 = DTO 215-F5 | Peanut cake, Kaduna, Nigeria | – | MG517682 | MG518053 | MG517873 | |
| DTO 087-A2 | Soil near road, Ifaty, Madagascar | MG662405 | MG517652 | MG517990 | MG517840 | |
| DTO 228-G1 = IBT 32079 | Stored rice grains from market, Abeokuta, Ogun State, Nigeria | MG662389 | MG517705 | MG518075 | MG517896 | |
| DTO 228-G3 = IBT 32086 = CBS 135587 | Sesame kernels from market, Plateau State, Vwrang, Nigeria | MG662387 | MG517707 | MG518077 | MG517898 | |
| DTO 228-G4 = IBT 32087 = CBS 135588 | Sesame kernels from market, Plateau State, Vwrang, Nigeria | – | MG517708 | MG518078 | MG517899 | |
| DTO 228-G5 = IBT 32088 = CBS 135589 | Sesame kernels from market, Plateau State, B/Ladi, Nigeria | – | MG517709 | MG518079 | MG517900 | |
| DTO 228-G6 = IBT 32089 = CBS 135404 | Sesame kernels from market, Plateau State, B/Ladi, Nigeria | – | MG517710 | MG518080 | MG517901 | |
| DTO 228-G7 = IBT 32090 = CBS 135405 | Sesame kernels from market, Plateau State, B/Ladi, Nigeria | – | MG517711 | MG518081 | MG517902 | |
| DTO 228-H2 = IBT 16807 | Mexican sesame seed imported to Denmark and sold in Lyngby, JC Frisvad, 1995 | – | MG517715 | MG518085 | MG517906 | |
| DTO 228-H3 = IBT 16808 | Mexican sesame seed imported to Denmark and sold in Lyngby, JC Frisvad, 1995 | – | MG517716 | MG518086 | MG517907 | |
| DTO 228-H7 = IBT 32083 | Agricultural soil, Minna State, Nigeria | – | MG517720 | MG518090 | MG517911 | |
| Aspergillus alliaceus | CBS 536.65NT = DTO 046-B1 = NRRL 315 = IMI 051982 = QM 1885 = ATCC 10760 = WB 315 = Thom 4656 = IBT 13377 = CCF 5607 | Dead blister beetle (Microbasis albida), Washington D.C., USA, M.M. High, neotype of Aspergillus alliaceus | EF661551 | EF661465 | EF661534 | MG517825 |
| CBS 110.26 = DTO 034-B2 = DTO 046-A7 = IBT 14351 = NRRL 316 = WB 316 = IMI 016125 = Thom 4660 = CCF 5603 | Allium cepa | MH279383 | MG517632 | MG518004 | MG517815 | |
| CBS 143682 = DTO 326-D5 = S757 = CCF 5416 = IBT 33356 | Intestine of Allolobophora hrabei, National Reservation Pouzdřanská step - Kolby, Czech Republic, A. Nováková, 2013 | MH279421 | MG517764 | MG518134 | MG517955 | |
| CBS 511.69 = DTO 368-C3 = IBT 13379 = CCF 5682 | Soil, Turkey | MH279439 | MG517786 | MG518156 | MG517976 | |
| CBS 542.65 = DTO 034-A9 = DTO 203-B1 = NRRL 4181 = ATCC 16891 = IBT 13378 = IMI 116711 = QM 1892 = WB 4181 = JH Warcup SA 117 | Soil, Australia, ex type of Petromyces alliaceus | EF661556 | EF661466 | EF661536 | EU021644 | |
| DTO 363-E8 = NRRL 318 = IBT 21073 = CCF 5601 | Unknown source | MH279430 | MG517776 | MG518146 | MG517967 | |
| DTO 363-E9 = IBT 23440 = EXF-670 = CCF 5605 | Saltern, Secovlje, Slovenia, P. Zalar | MH279431 | MG517777 | MG518147 | MG517968 | |
| DTO 363-F1 = IBT 21992 = A196 = CCF 5604 | Mixed feed, Spain | MH279432 | MG517778 | MG518148 | MG517969 | |
| DTO 363-F2 = IBT 21754 = IMI 017295 = CCF 5606 | Contaminant in ex type culture of Aspergillus wentii | MH279433 | MG517779 | MG518149 | – | |
| DTO 368-C4 = IMI 226007 = IBT 14130 = CCF 5680 | Soil, Calicut University, India | MH279440 | MG517787 | MG518157 | MG517977 | |
| IBT 21770 | Prairie soil, Nebraska, USA | MH279446 | MG517790 | MG518161 | MG517980 | |
| Mo2 | Soil above the Movile cave, Romania, 2011, A. Nováková | MH279443 | MG517791 | LT558734 | MG517981 | |
| NRRL 1206 = Thom 5741 | Unknown source | EF661543 | EF661463 | EF661535 | EU021622 | |
| NRRL 20602 = ATCC 58745 = IBT 14317 = UAMH 2476 | Clinical isolate from human ear, Alberta, Canada, ex type of Aspergillus albertensis | EF661548 | EF661464 | EF661537 | EU021628 | |
| S862 = CCF 4954 | Soil above the Movile cave, Romania, A. Nováková, 2013 | MH279444 | MG517615 | MG518160 | MG517795 | |
| S916 = CCF 5434 | Allolobophora hrabei casts, National Monument Ječmeniště, Czech Republic, A. Nováková, 2013 | MH279442 | MG517616 | MG518159 | MG517796 | |
| S98 = CCF 4953 | Soil above Movile cave, Romania, A. Nováková, 2012 | MH279445 | MG517614 | LT558735 | MG517794 | |
| Aspergillus arachidicola | CBS 117610T = DTO 009-G3 = IBT 25020 | Arachis glabrata leaf, Mercedes, Corrientes province, Argentina, ex type of Aspergillus arachidicola | MF668184 | EF203158 | EF202049 | MG517802 |
| CBS 117611 = DTO 009-G4 = IBT 27185 | Arachis glabrata leaf, Mercedes, Corrientes province, Argentina | - | MG517620 | MG518006 | MG517803 | |
| CBS 117615 = DTO 010-H5 = IBT 28178 | Arachis glabrata leaf, Ituzaingó, Corrientes province, Argentina | – | MG517627 | MG517999 | MG517810 | |
| DTO 228-H9 | Leaf of Protea roupelliae var. roupelliae, Buffelskloof, South Africa | MG662384 | MG517721 | MG518091 | MG517912 | |
| Aspergillus aspearensis | CBS 143672T = DTO 203-D9 = CCTU758 = IBT 32590 = IBT 34544 | Soil, Aspear Island, Urmia Lake, Iran, soil, ex type of Aspergillus aspearensis | MG662398 | MG517669 | MG518040 | MG517857 |
| DTO 203-D4 = CBS 143671 = CCTU753 = IBT 34543 | Soil, Aspear Island, Urmia Lake, Iran | MG662399 | MG517667 | MG518038 | MG517855 | |
| DTO 203-E1 = CBS 143673 = CCTU759 = IBT 32591 | Soil, Aspear Island, Urmia Lake Iran | MH279394 | MG517670 | MG518041 | MG517858 | |
| Aspergillus austwickii | CBS 143677T = DTO 228-F7 = IBT 32590 = IBT 32076 | Stored rice grains from market, Abeokuta, Ogun State, Nigeria, ex type of Aspergillus austwickii | MG662391 | MG517702 | MG518072 | MG517893 |
| CBS 135406 = DTO 228-G8 = IBT 32091 | Sesame kernels from market, Plateau State, B/Ladi, Nigeria | MG662386 | MG517712 | MG518082 | MG517903 | |
| CBS 143678 = DTO 228-F8 = IBT 32077 | Stored rice grains from market, Abeokuta, Ogun State, Nigeria | – | MG517703 | MG518073 | MG517894 | |
| DTO 228-F9 = IBT 32078 | Stored rice grains from market, Abeokuta, Ogun State, Nigeria | MG662390 | MG517704 | MG518074 | MG517895 | |
| Aspergillus avenaceus | CBS 109.46T = DTO 009-H6 = DTO 006-A2 = NRRL 517 = ATCC 16861 = IMI 016140 = LCP 89.2592 = LSHB BB 155 = QM 6741 = WB 317 = IBT 4376 = IBT 4555 | Green pea (Pisum sativum), United Kingdom, G.E. Turfitt, 1938, ex type of Aspergillus avenaceus | AF104446 | FJ491481 | FJ491496 | JN121424 |
| CBS 102.45 = NCTC 6548 | Unknown source, United Kingdom | – | FJ491480 | FJ491495 | – | |
| Aspergillus bertholletius | CBS 143687 = DTO 223-D3 = IBT 29228 = CCT 7615T = ITAL 270/06 | Soil close to Bertholletia excelsa trees, Amazonian rainforest, Brazil, ex type of Aspergillus betholletius | JX198673 | MG517689 | JX198674 | MG517880 |
| IBT 29227 = ITAL 275/06 = CCT 7618 | Soil close to Bertholletia excelsa trees, Amazonian rainforest | – | – | – | – | |
| IBT 30617 = ITAL 272/06 = CCT 7617 | Soil close to Bertholletia excelsa trees, Amazonian rainforest | – | – | – | – | |
| DTO 223-D4 = IBT 30618 = ITAL 271/06 = CCT 7616 | Soil close to Bertholletia excelsa trees, Amazonian rainforest | – | – | – | – | |
| IBT 31739 = ITAL 262 = CCT 7614 | Bertholletia excelsa nut shell, Market, Amazon | – | – | – | – | |
| Aspergillus caelatus | CBS 763.97T = DTO 046-A8 = NRRL 25528 = IBT 21091 | Soil, USA, ex type of Aspergillus caelatus | AF004930 | MG517640 | MG518018 | MG517823 |
| CBS 764.97 = NRRL 25404 | Soil, USA | – | EF203129 | EF202036 | - | |
| DTO 276-I2 | Corn silage, Cordoba, Argentina | – | MG517738 | MG518108 | MG517929 | |
| DTO 285-H9 | Soil from corn field, Thailand | – | MG517751 | MG518121 | MG517942 | |
| DTO 285-I1 | Soil from corn field, Thailand | – | MG517752 | MG518122 | MG517943 | |
| NRRL 25566 = IBT 29770 = DTO 073-B7 | Soil, Japan | – | MG517651 | MG518025 | MG517839 | |
| NRRL 25567 = IBT 29773 = DTO 073-B8 | Soil, Japan | – | – | – | – | |
| NRRL 25568 = IBT 29772 = DTO 073-B9 | Soil, Japan | – | – | – | – | |
| NRRL 25569 = IBT 29771 = DTO 073-C1 | Soil, Japan | – | – | – | – | |
| NRRL 26100 | Soil of peanut field, 2.5 km east of Herod, Georgia, USA | EF661550 | EF661471 | EF661523 | EF661437 | |
| Aspergillus cerealis | CBS 143674T = DTO 228-E7 = IBT 32067 | Stored rice grains from market, Shagamu, Ogun State, Nigeria, ex type of Aspergillus cerealis | MG662394 | MG517693 | MG518063 | MG517884 |
| CBS 143675 = DTO 228-E8 = IBT 32068 | Stored rice grains from market, Shagamu, Ogun State, Nigeria | – | MG517694 | MG518064 | MG517885 | |
| CBS 143676 = DTO 228-E9 = IBT 32069 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | MG662393 | MG517695 | MG518065 | MG517886 | |
| DTO 228-E6 = IBT 32076 | Stored rice grains from market, Shagamu, Ogun State, Nigeria | MG662395 | MG517692 | MG518062 | MG517883 | |
| DTO 228-F1 = IBT 32070 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | MG662392 | MG517696 | MG518066 | MG517887 | |
| DTO 228-F2 = IBT 32071 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | – | MG517697 | MG518067 | MG517888 | |
| DTO 228-F3 = IBT 32072 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | – | MG517698 | MG518068 | MG517889 | |
| DTO 228-F4 = IBT 32073 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | – | MG517699 | MG518069 | MG517890 | |
| DTO 228-F5 = IBT 32074 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | – | MG517700 | MG518070 | MG517891 | |
| DTO 228-F6 = IBT 32075 | Stored maize grains from market, Shagamu, Ogun State, Nigeria | – | MG517701 | MG518071 | MG517892 | |
| MACI219 = NRRL 66709 | Peanut pods, Pokaha, Karhogo region, North part of Côte d’Ivoire (Ivory Coast) | KY689208 | KY628791 | KY661266 | – | |
| MACI254 = NRRL 66710 | Peanut pods, Gbandokaha, Karhogo region, North part of Côte d’Ivoire, 2014, probably ex type of Aspergillus korhogoensis | KY689209 | KY628792 | KY661267 | – | |
| MACI264 = NRRL 66711 | Peanut pods, Gbandokaha, Karhogo region, North part of Côte d’Ivoire, 2014 | KY689210 | KY628793 | KY661268 | – | |
| MACI46 = NRRL 66708 | Peanut pods, Karhogo region, North part of Côte d’Ivoire, 2014 | KY689207 | KY628790 | KY661265 | – | |
| Aspergillus coremiiformis | CBS 553.77T = DTO 046-A3 = ATCC 38576 = IHEM 4503 = IMI 223069 = NRRL 13603 = NRRL 13756 = IBT 3822 = IBT 13506 = IBT 21944 | Soil, Tai National Forest, Ivory Coast, ex type of Aspergillus coremiiformis | AJ874114 | FJ491482 | FJ491488 | JN121533 |
| Aspergillus flavus | CBS 100927T = NRRL 1957 = ATCC 16883 = CBS 569.65 = IMI 124930 = IBT 3605 = IBT 3610 | Cellophane diaphragm of an optical mask, South Pacific Islands, ex type of Aspergillus flavus | AF027863 | EF661485 | EF661508 | EF661440 |
| AF70 | Seed of upland cotton (Gossypium hirsutum), Arizona, USA, genome sequenced | – | genome* | genome* | genome* | |
| CBS 110.55 = DTO 046-A1 = ATCC 12073 = NRRL 4743 = IMUR 236 = QM 6951 = WB 4743 = IBT 3819 | Air contaminant, Brazil, ex type of Aspergillus fasciculatus | FJ491463 | EF203135 | MG518005 | MG517821 | |
| CBS 117637 = DTO 009-F9 = IBT 27177 | Arachis hypogea seed, Provincia de Formosa, Las Lomitas, Argentina | – | MG517618 | MG518010 | MG517800 | |
| CBS 117638 = DTO 009-G1 | Arachis hypogea seed, Provincia de Corrientes, Empedrado, Argentina | – | MG517619 | MG518011 | MG517801 | |
| CBS 117732 = NRRL 3251 = IBT 3597 = IBT 3618 | Walnut, USA (small sclerotia) | – | – | – | – | |
| CBS 118.62 = DTO 010-H6 = IFO 7600 = IMI 091548 = NRRL A-11608 = RIB 1406 | Arachis hypogea, Brazil | – | MG517628 | MG517996 | MG517811 | |
| CBS 119368 = DTO 011-I2 = KACC 41730 | Wheat, Boun-up, Boukun, Chungbuk Prov., South Korea | – | MG517630 | MG518002 | MG517813 | |
| CBS 120.51 = DTO 046-A4 = ATCC 16859 = IFO 8135 = IMI 045644 = LCP 56.1517 = LSHB BB213 = NRRL 2097 = NRRL A-2022 = QM 6871 = WB 2097 = IBT 3636 | Culture contaminant, London, England, ex type of Aspergillus thomii | EF661549 | MG517639 | MG518012 | MG517822 | |
| CBS 128202 = NRRL 3357 = ATCC 200026 = IBT 3696 = IBT 28518 = IBT 29624 | Peanut cotyledons, USA, genome sequenced | JX535495 | genome* | genome* | genome* | |
| CBS 133263 = DTO 215-E9 | Edible mushroom from market, Lagos State, Nigeria | – | JX627689 | JX627693 | MG517868 | |
| CBS 143688 = DTO 359-D8 = KACC 46894 = IBT 34547 | Air, South Korea | – | MG517774 | MG518144 | MG517965 | |
| CBS 143689 = DTO 359-D9 = KACC 46895 = IBT 34548 | Air, South Korea | – | MG517775 | MG518145 | MG517966 | |
| CBS 485.65 = DTO 046-B7 = ATCC 16870 = IFO 5324 = IMI 124932 = LCP 89.3556 = NRRL 4818 = WB 4818 = IBT 3641 = IBT 3657 | Butter, Japan, ex type of Aspergillus flavus var. columnaris and A. flavus var. asper | EF661563 | MG517643 | MG518014 | MG517828 | |
| CBS 501.65 = DTO 046-B5 = ATCC 16862 = IMI 044882 = NRRL 4998 = WB 4998 = IBT 4378 = IBT 4402 | Cotton lintafelt, England, ex neotype of Aspergillus subolivaceus | EF661563 | MG517642 | MG518015 | MG517827 | |
| CBS 542.69 = DTO 046-B4 = IMI 141553 = NRRL 3751 = GKC 1421(1) = IBT 3649 | Stratigraphic core sample, soil, Niigata Pref., Kambara, Japan, ex type of Aspergillus kambarensis | EF661554 | MG517641 | MG518016 | MG517826 | |
| CBS 574.65 = DTO 303-C3 = ATCC 1010 = IMI 016142 = IMI 124935 = NRRL 506 = NRRL 1653 | Corn (Zea mays), Vermont, USA, representative of A. effusus fideThom & Church (1926) and Thom & Raper (1945) (Raper & Fennell 1965:377) | JN185448 | JN185446 | JN185447 | JN185449 | |
| DTO 016-I5 = dH 16719 | Infection of leg (after liver transplantation), male 43 year old, China | – | MG517631 | MG518003 | MG517814 | |
| DTO 062-C7 | Peanut, Indonesia | – | MG517645 | MG517985 | MG517832 | |
| DTO 062-C8 | Peanut, Indonesia | – | MG517646 | MG517986 | MG517833 | |
| DTO 062-H7 | Peanut, Indonesia | – | MG517647 | MG517987 | MG517834 | |
| DTO 066-C3 | Corn kernels, Indonesia | – | MG517650 | MG517989 | MG517837 | |
| DTO 087-A3 | Forest soil, Ifaty, Madagascar | – | MG517653 | MG517991 | MG517841 | |
| DTO 087-A4 | Forest soil, Ifaty, Madagascar | – | MG517654 | MG517992 | MG517842 | |
| DTO 215-E5 | Laboratory contaminant, Nigeria | – | MG517679 | MG518050 | MG517867 | |
| DTO 258-C9 | Corn kernels, from East.Europe, imported to the Netherlands | – | MG517725 | MG518095 | MG517916 | |
| DTO 258-D6 | Corn kernels, from East.Europe, imported to the Netherlands | – | MG517728 | MG518098 | MG517919 | |
| DTO 276-H7 | Poultry feedstuff, Cordoba, Argentina | – | MG517734 | MG518104 | MG517925 | |
| DTO 276-H8 | Poultry feedstuff, Cordoba, Argentina | – | MG517735 | MG518105 | MG517926 | |
| DTO 276-H9 | Poultry feedstuff, Cordoba, Argentina | – | MG517736 | MG518106 | MG517927 | |
| DTO 276-I1 | Poultry feedstuff, Cordoba, Argentina | – | MG517737 | MG518107 | MG517928 | |
| DTO 276-I3 | Corn silage, Cordoba, Argentina | – | MG517739 | MG518109 | MG517930 | |
| DTO 276-I4 | Chinchilla feedstuffs, Cordoba, Argentina | – | MG517740 | MG518110 | MG517931 | |
| DTO 276-I5 | Chinchilla feedstuffs, Cordoba, Argentina | – | MG517741 | MG518111 | MG517932 | |
| DTO 276-I6 | Chinchilla feedstuffs, Cordoba, Argentina | – | MG517742 | MG518112 | MG517933 | |
| DTO 276-I7 | Chinchilla feedstuffs, Cordoba, Argentina | – | MG517743 | MG518113 | MG517934 | |
| DTO 276-I8 | Chinchilla feedstuffs, Cordoba, Argentina | – | MG517744 | MG518114 | MG517935 | |
| DTO 281-E2 | Rice, Thailand | – | MG517745 | MG518115 | MG517936 | |
| DTO 281-H8 | Rice, Thailand | – | MG517746 | MG518116 | MG517937 | |
| DTO 285-F6 | Soil from corn-field, Thailand | – | MG517748 | MG518118 | MG517939 | |
| DTO 285-G3 | Soil from corn-field, Thailand | – | MG517749 | MG518119 | MG517940 | |
| DTO 285-I4 | Soil from corn-field, Thailand | – | MG517753 | MG518123 | MG517944 | |
| DTO 300-C7 | Corn kernels, imported into the Netherlands | – | MG517754 | MG518124 | MG517945 | |
| DTO 300-D7 | Corn kernels, imported into the Netherlands | – | MG517755 | MG518125 | MG517946 | |
| DTO 359-D7 = IBT 34546 = KACC 46893 | Air, South Korea | – | – | – | – | |
| DTO 359-E1 = IBT 34551 = KACC 46897 | Corn, South Korea | – | – | – | – | |
| DTO 359-E2 = IBT 34550 = KACC 46913 | Soil, South Korea | – | – | – | – | |
| DTO 359-E3 = KACC 46917 | Soil, Gyoenggi, Suwon, Korea | |||||
| NRRL 20521 | Corn, Mississippi, USA | EF661547 | EF661492 | EF661514 | EF661447 | |
| NRRL 3518 = NRRL A-14304 | Wheat flour, Peoria, Illinois, USA | EF661552 | EF661487 | EF661510 | EF661442 | |
| NRRL 4822 | Unknown source | EF661564 | EF661490 | EF661513 | EF661445 | |
| Aspergillus hancockii | FRR 3425T = CBS 142004 = DTO 360-G7 | Cultivated soil, Queensland, Australia, ex type of Aspergillus hancockii | KX858342 | MBFL01001228.1:26000-28000 | MBFL01000377.1:5000-7000 | MBFL01000137:9000-11000 |
| CBS 142001 = FRR 5050 = DTO 360-G4 = IBT 35030 | Soil, Lockhart, New South Wales, Australia, J.I. Pitt, 2003 | – | – | – | – | |
| CBS 142002 = FRR 6103 = DTO 360-G5 = IBT 35031 | Dried peas, Victoria, Australia, M. Bull, 1997 | – | – | – | – | |
| Aspergillus lanosus | CBS 650.74T = IMI 130727 = QM 9183 = IBT 33634 | Soil under Tectona grandis, Uttar Pradesh, India | FJ491471 | MG517633 | MG518017 | EU021642 |
| Aspergillus leporis | CBS 151.66T = IBT 3609 = DTO 199-B2 = CBS 129302 = RMF 99 = WB 5188 = ATCC 16490 = LCP 89.2583 = NRRL 3216 | Dung of Lepus townsensii, near Saratoga, Wyoming, USA, ex type of Aspergillus leporis | MH279391 | MG517662 | MG518033 | MG517850 |
| CBS 125914 = DTO 195-C3 = R1251 | A1 horizon soil, open area in sagebrush grassland, Rock Springs, Wyoming, USA (DOE site, 11 km west of Rock Springs) | MH279389 | MG517660 | MG518031 | MG517848 | |
| CBS 129235 = DTO 303-C5 | Plant root tissue at non-seleniferous soil, Nunn, Colorado, USA | – | MG517760 | MG518130 | MG517951 | |
| CBS 129310 = RMF 9587 = DTO 201-H1 | A1 horizon soil, Canyonlands National Park, Utah, USA | MH279392 | MG517663 | MG518034 | MG517851 | |
| CBS 129330 = RMF 7757 = DTO 202-A2 | Soil beneath Atriplex confertifolia, near Jim Bridger Power Plant, Sweetwater County, Wyoming, USA | MH279393 | MG517664 | MG518035 | MG517852 | |
| CBS 129596 = DTO 206-A8 = RMF G74 | A1 horizon soil from bunchgrass rhizosphere, sagebrush grassland, Rock Springs, Wyoming, USA | MH279395 | MG517673 | MG518044 | MG517861 | |
| CBS 132153 = DTO 210-E1 | Surface soil, near Dubois, Wyoming, USA | MH279396 | MG517674 | MG518045 | MG517862 | |
| CBS 132177 = RMF 2050 = DTO 210-G5 | A1 Horizon soil, Grand Teton National Park, Wyoming, USA | MH279397 | MG517676 | MG518047 | MG517864 | |
| CBS 349.81 = IBT 3600 = NRRL 6599 = DTO 303-C4 = ATCC 44565 = Strain O168 | Soil, Wyoming, USA | EF661569 | EF661500 | EF661542 | EF661460 | |
| IBT 12296 = IBT 13578 = ATCC 76617 | Soil undre grass, Canyon de Chelly, Arizona, USA | – | – | – | – | |
| IBT 16309 = RMF A39 | Soil under Atriplex gardneri, cool desert, 10 km north of Rock Springs, Great Divide Basin, Wyoming, USA | – | – | – | – | |
| IBT 16585 | Soil under Atriplex confertifolia, cool desert, 10 km north of Rock Springs, Great Divide Basin, Wyoming, USA | – | – | – | – | |
| CBS 132178 = RMF 2110 = DTO 210-G6 | A1 Horizon soil, Grand Teton National Park, Wyoming, USA | MH279398 | MG517677 | MG518048 | MG517865 | |
| Aspergillus luteovirescens | CBS 620.95T = DTO 010-H1 | Unknown source, ex type of Aspergillus luteovirescens | MG662406 | MG517625 | MG517998 | MG517808 |
| CBS 117187 = NRRL 25010 = IBT 23536 | Frass in a sílkworm rearing house, Japan, 1987, ex type of Aspergillus bombycis | AF104444 | EF661498 | EF661533 | EF661458 | |
| DTO 073-C3 = NRRL 29236 = IBT 29777 | Frass in a silkworm rearing house, 1983, Ibaraki Prefecture, Japan | – | – | – | – | |
| DTO 073-C4 = NRRL 29237 = IBT 29780 | Frass in a silkworm rearing house, 1983, Ibaraki Prefecture, Japan | – | – | – | – | |
| DTO 073-C5 = NRRL 29241 = IBT 29779 | Frass in a silkworm rearing house, 1983, Oita Prefecture, Japan | – | – | – | – | |
| ITAL 246 = IBT 31534 | Brazil nut, Amazon, Brazil | – | – | – | – | |
| NRRL 25593 = IBT 23535 | Frass in a sílkworm rearing house, Japan, 1987 | AF104445 | EF661497 | EF661532 | EF661457 | |
| NRRL 29235 = DTO 073-C2 = IBT 23537 = IBT 29778 | Frass in a silkworn rearing house, Indonesia, 1999 | AF338641 | AY017575 | AY017622 | – | |
| Aspergillus minisclerotigenes | CBS 117635T = DTO 009-F7 = IBT 25032 | Arachis hypogea, Manfredi, Córdoba province, Argentina, ex type of Aspergillus minisclerotigenes | EF409239 | EF203148 | MG518009 | MG517799 |
| CBS 117633 = DTO 009-F5 | Arachis hypogea seed, Provincia de Formosa, Las Lomitas, Argentina | MG662408 | EF203153 | MG518007 | MG517797 | |
| CBS 117634 = DTO 009-F6 = IBT 27197 | Arachis hypogea seed, Provincia de Cordoba, Alejandro, Argentina | MG662402 | MG517617 | MG518008 | MG517798 | |
| DTO 045-F4 = FRR 4086 | Freshly pulled peanuts, Interlaw Road, Kingaropy, Queensland, Australia | – | MG517635 | MG518021 | MG517817 | |
| DTO 045-F5 = FRR 4937 | Soil, Australia | – | MG517636 | MG518022 | MG517818 | |
| DTO 045-F6 = FRR 5309 | Soil, Coalston Lakes, Queensland, Australia | – | MG517637 | MG518023 | MG517819 | |
| DTO 045-I9 = NRRL A-11611 = NRRL 6444 = IBT 3840 | Soil of peanut field, Nigeria | MH279386 | MG517638 | MG518024 | MG517820 | |
| DTO 228-G9 = IBT 32094 | Agricultural soil, Jos, Plateau State, Nigeria | – | MG517713 | MG518083 | MG517904 | |
| DTO 228-H1 = IBT 32111 | Agricultural soil, Minna, Niger State, Nigeria | – | MG517714 | MG518084 | MG517905 | |
| DTO 228-H5 = IBT 24629 | Curry powder from Kenya imported to Denmark | – | MG517718 | MG518088 | MG517909 | |
| Aspergillus mottae | CBS 130016T = DTO 223-C8 = IBT 32309 = MUM 10.231 | Maize kernel, Braga, Portugal, ex type of Aspergillus mottae | JF412767 | MG517687 | MG518058 | MG517878 |
| MUM 10.233 | Maize, Portugal | – | HM803090 | HM803013 | HM802982 | |
| Aspergillus neoalliaceus | CBS 143681T = DTO 326-D3 = S765 = CCF 5433 = IBT 33110 = IBT 33353 | Soil, Czech Republic, National Reservation Pouzdřanská step - Kolby, A. Nováková, 2013, ex type of Aspergillus neoalliaceus | MH279420 | MG517763 | MG518133 | MG517954 |
| CBS 134375 = S77 = CCF 4424 | Soil, National Monument Ječmeniště, Czech Republic, A. Nováková, 2012 | MH279441 | MG517613 | MG518158 | MG517793 | |
| DTO 326-D6 = S768 = CCF 5414 = IBT 33111 = IBT 33357 | Drilosphere soil, National Reservation Pouzdřanská step – Kolby, Czech Republic, A. Nováková, 2013 | MH279422 | MG517765 | MG518135 | MG517956 | |
| DTO 326-D7 = B6 = CCF 5408 = IBT 32726 | Soil, National Reservation Pouzdřanská step – Kolby, Czech Republic, A. Nováková, 2010 | MH279423 | MG517766 | MG518136 | MG517957 | |
| DTO 326-E1 = S756 = CCF 5410 = IBT 33359 | Soil, National monument Ječmeniště, Czech Republic, A. Nováková, 2013 | MH279424 | MG517768 | MG518138 | MG517959 | |
| DTO 326-E2 = S766 = CCF 5412 = IBT 33355 | Allolobophora hrabei cast, National Reservation Pouzdřanská step – Kolby, Czech Republic, A. Nováková, 2013 | MH279425 | MG517769 | MG518139 | MG517960 | |
| DTO 326-E4 = S764 = CCF 5411 = IBT 33358 | Soil, National monument Ječmeniště, Czech Republic, A. Nováková, 2013 | MH279426 | MG517770 | MG518140 | MG517961 | |
| DTO 326-E5 = S913 = CCF 5415 = IBT 33351 | Soil, National monument Ječmeniště, Czech Republic, A. Nováková, 2013 | MH279427 | MG517771 | MG518141 | MG517962 | |
| DTO 326-E7 = S767 = CCF 5413 = IBT 33109 = IBT 33352 | Soil, National Reservation Pouzdřanská step – Kolby, Czech Republic, A. Nováková, 2013 | MH279428 | MG517772 | MG518142 | MG517963 | |
| CCF 5815 = S1429 | Soil, above the Liliecilor de la Gura Dobrogei cave, Dobrogea, Romania, A. Nováková, 2016 | – | – | – | – | |
| CCF 5840 = S988 | Soil, above the Limanu cave, Dobrogea, Romania, A. Nováková, 2014 | – | – | – | – | |
| Aspergillus nomius | CBS 260.88T = NRRL 13137 = IBT 3656 = IBT4966 = FDA M93 | Wheat, USA, A.F. Schindler, 1965, ex type of Aspergillus nomius | AF027860 | EF661494 | EF661531 | EF661456 |
| CBS 117629 = NRRL 25585 = IBT 23530 | Silk worm frass, Japan, 1987 | – | – | – | – | |
| CBS 399.93 = DTO 301-I8 = AS 3.4626 = IBT 14647 | Soil, Guandong, Zhaoqing, China, ex type of Aspergillus zhaoqingensis | FJ491472 | MG517757 | MG518127 | MG517948 | |
| DTO 161-F1 | Bamboo sample, Walailak, Thailand | MH279387 | MG517656 | MG518026 | MG517844 | |
| DTO 161-F2 | Bamboo sample, Addis Abeba, Ethiopia | MH279388 | MG517657 | MG518027 | MG517845 | |
| DTO 226-I5 | Storage room of cassava, Yogyakarta, Indonesia | – | MG517690 | MG518060 | MG517881 | |
| DTO 227-B8 | Storage room of cassava, Yogyakarta, Indonesia | – | MG517691 | MG518061 | MG517882 | |
| DTO 243-E8 | HIV-Care room, Indonesia | – | MG517722 | MG518092 | MG517913 | |
| DTO 247-F9 | House dust, Mexico | – | MG517723 | MG518093 | MG517914 | |
| DTO 247-G8 | House dust, Mexico | – | MG517724 | MG518094 | MG517915 | |
| DTO 318-F4 | Heat treated pectin, Germany | – | MG517761 | MG518131 | MG517952 | |
| DTO 321-F2 | Cystic fibrosis patient material, the Netherlands | MH279419 | MG517762 | MG518132 | MG517953 | |
| IMI 190557 = NRRL 20745 = IBT 19368 | Dried Curcuma longa, Central Crops Reseaerch Institute, India | AF338612 | AY017543 | AY017590 | – | |
| NRRL 13138 = IBT 4493 = IBT 4495 = IBT 5054 | Sub-isolate from a mixed culture, U.L. Diener, 1967 | – | – | – | – | |
| NRRL 3161 = IBT 3661 = IBT 4975 | Cycas circinalis, Guam, USA, A.C. Keyl, 1965 | AF338642 | EF661493 | EF661530 | EF661453 | |
| Aspergillus novoparasiticus | CBS 126849T = DTO 223-C3 = DTO 223-C4 = FMR 10121 = LEMI 250 = IBT 32311 | Sputum of leukemic patient, Sao Paolo, Brazil, ex type of Aspergillus novoparasiticus | MG662397 | MG517684 | MG518055 | MG517875 |
| CBS 126850 = DTO 223-C5 = FMR 10158 = LEMI 149 IOP = IBT 32312 | Air sample, Sao Paulo, Brazil | MH279415 | MG517686 | MG518057 | MG517877 | |
| Aspergillus oryzae | CBS 102.07T = CBS 110.47 = CBS 100925 = ATCC 1011 = ATCC 12891 = ATCC 4814 = ATCC 7651 = ATCC 9102 = CECT 2094 = IFO 4075 = IFO 5375 = IMI 016266ii = IMI 016266 = IMI 044242 = LSHBA c.19 = NCTC 598 = NRRL 447 = NRRL 692 = QM 6735 = Thom 113 = WB 447 = IBT 21451 | Unknown source, ex type of Aspergillus oryzae | EF661560 | EF661483 | EF661506 | EF661438 |
| NRRL 458 = ATCC 10063 = ATCC 9376 = IMI 051983 | Unknown source | EF661562 | EF661484 | EF661507 | EF661439 | |
| RIB40 = ATCC 42149 = JCM 13832 = NRRL 5590 = IBT 28103 | Horsebean, Muruka soy saúce factory, Mimaki-mura, Kuse-gun, Kyoto, Japan, genome sequenced | – | genome* | genome* | genome* | |
| Strain 100-8 | Mutant of A. oryzae 3.042, which is used in soy sauce fermentation, China, genome sequenced | – | genome* | genome* | genome* | |
| Aspergillus parasiticus | CBS 100926T = NRRL 502 = ATCC 1018 = ATCC 6474 = ATCC 7865 = IMI 015957 = IMI 015957ii = IMI 015597iv = IMI 015957vi = IMI 015957vii = IMI 015957ix = NRRL 1731 = IBT 3607 | Sugar cane mealy bug (Pseudococcus calceolariae), Hawaii, USA, ex neotype of Aspergillus parasiticus | AF027862 | EF661481 | EF661516 | EF661449 |
| CBS 104.22 = DTO 009-H2 = IFO 5867 | Unknown source | – | MG517621 | MG517994 | MG517804 | |
| CBS 119.51 = DTO 009-H3 = IFO 5337 | Unknown substrate, Japan | – | MG517622 | MG518000 | MG517805 | |
| CBS 138.52 = DTO 009-H4 | Unknown substrate, Japan | – | MG517623 | MG517997 | MG517806 | |
| CBS 260.67 = DTO 046-C2 = ATCC 15517 = CCM F-550 = CECT 2680 = DSM 2038 = IFO 30179 = IHEM 4387 = IMI 120920 = IMI 229041 = MUCL 31311 | Unknown source, Japan, ex type of Aspergillus parasiticus var. globosus | MG662400 | EF203156 | MG518013 | MG517830 | |
| CBS 580.65 = DTO 046-B9 = ATCC 1014 = ATCC 16863 = IMI 016127ii = LSHB Ac22 = NCTC 974 = NRRL 424 = QM 7475 = VKM F-2041 = WB 424 = IBT 3664 = IBT 3670 = IBT 10828 | Soil, Georgia, USA, ex type of Aspergillus terricola var. americana | MG662404 | MG517644 | MG518030 | MG517829 | |
| CBS 822.72 = DTO 046-A9 = ATCC 22789 = IFO 30109 = IMI 089717 = RIB 4002 = TRI M 39 = IBT 4377 = IBT 4408 | Arachis hypogea, Uganda, ex type of Aspergillus toxicarius | MG662401 | EF203163 | MG518019 | MG517824 | |
| CBS 921.70 = ATCC 26691 = CECT 2681 = IHEM 4383 = NRRL 2999 = IBT 3634 = IBT 15675 | Unknown source, Uganda | AB008418 | – | – | – | |
| DTO 203-C4 | Soil, Aspear Island, Iran | – | MG517666 | MG518037 | MG517854 | |
| DTO 203-H7 | Soil, Kabodan Island, Iran | – | MG517672 | MG518043 | MG517860 | |
| DTO 258-D1 | Corn kernels from East-Europe imported to the Netherlands | – | MG517726 | MG518096 | MG517917 | |
| DTO 258-D4 | Corn kernels from East-Europe imported to the Netherlands | – | MG517727 | MG518097 | MG517918 | |
| DTO 283-C6 | Soil from corn.field, Thailand | – | MG517747 | MG518117 | MG517938 | |
| DTO 285-G9 | Soil from corn.field, Thailand | – | MG517750 | MG518120 | MG517941 | |
| DTO 301-E6 | Corn kernels, imported to the Netherlands | – | MG517756 | MG518126 | MG517947 | |
| DTO 303-C2 | Unknown source | – | MG517759 | MG518129 | MG517950 | |
| NRRL 13005 = IBT 4564 | Microarthropod in beech forest litter, Michigan, USA (produces sclerotia) | – | – | – | – | |
| NRRL 4123 | Toxic grain | EF661555 | EF661479 | EF661518 | EF661451 | |
| NRRL 6433 = IBT 4375 | Corn, North Carolina, USA | EF661568 | EF661480 | EF661519 | EF661452 | |
| Aspergillus pipericola | CBS 143680T = DTO 228-H4 = IBT 24628 | Black pepper, unknown origin, imported to Denmark, ex type of Aspergillus pipericola | MG662385 | MG517717 | MG518087 | MG517908 |
| Aspergillus pseudocaelatus | CBS 117616T = DTO 010-H4 = IBT 27191 | Arachis burkartii leaf, Mercedes, Corrientes province, Ituzaingó, Argentina | EF409242 | MG517626 | MG517995 | MG517809 |
| ITAL 103CC = IBT 29230 | Peanuts, Brazil | – | – | – | – | |
| ITAL 1300F/09 = IBT 30532 | Brazil nuts, Amazon, Brazil | – | – | – | – | |
| Aspergillus pseudonomius | CBS 119388T = DTO 009-F1 = NRRL 3353 = IBT 27864 = IBT 14897 | Diseased alkali bee (Nomius sp.), Wyoming, USA | AF338643 | EF661495 | EF661529 | EF661454 |
| DTO 177-G7 | Soil of corn-field, Phayao, Thailand | – | MG517659 | MG518029 | MG517847 | |
| DTO 262-F3 | Indoor environment of child hospital, Izmir, Turkey | – | MG517729 | MG518099 | MG517920 | |
| DTO 267-D6 | House dust, Micronesia | MH279416 | MG517731 | MG518101 | MG517922 | |
| DTO 267-H7 | House dust, Thailand | MH279417 | MG517732 | MG518102 | MG517923 | |
| DTO 267-I4 | House dust, Thailand | – | MG517733 | MG518103 | MG517924 | |
| IBT 12657 = DTO 303-A4 | Seed, unknown location | MH279418 | MG517758 | MG518128 | MG517949 | |
| ITAL 823/07 | Brazil nut, Amazon, Brazil | – | – | – | – | |
| ITAL 849F = IBT 32759 | Brazil nut, Amazonas, Brazil | – | – | – | – | |
| NRRL 6552 | Diseased pine sawfly, Wisconsin, USA, C.R. Benjamin, 1967 | – | EF661496 | EF661528 | EF661455 | |
| Aspergillus pseudotamarii | CBS 766.97T = NRRL 25517 = DTO 046-C1 = IBT 21092 | Soil, teafield, Japan | AF272574 | EF661477 | EF661521 | EU021631 |
| CBS 117625 = NRRL 25518 = IBT 21090 | Soil, teafield, Japan | – | – | – | – | |
| CBS 117628 = NRRL 25519 = IBT 21093 | Soil, teafield, Japan | – | – | – | – | |
| CBS 765.97 = NRRL 443 | Unknown source | AF004931 | EF661476 | EF661520 | EU021650 | |
| ITAL 791F/09 = IBT 30530 | Brazil nut, Amazonas, Brazil | – | – | – | – | |
| ITAL 792F/09 = IBT 30531 | Brazil nut, Amazonas, Brazil | – | – | – | – | |
| Aspergillus sergii | CBS 130017T = DTO 223-C9 = IBT 32292 = IBT 32293 | Fruits of Prunus dulcis, Trans-Os-Montes processing plant, Faro, Portugal, ex type of Aspergillus sergii | JF412769 | MG517688 | MG518059 | MG517879 |
| Aspergillus sojae | CBS 100928T = DTO 046-C3 = ATCC 42251 = IAM 2669 = IFO 4244 = IFO 30112 = IMI 191300 = RIB 1045 = SRRC 1126 = K. Sakaguchi SH-10-6 = IBT 21642 = IBT 32109 | Koji of soy sauce, shoyu brewing, 1942, ex neotype of Aspergillus sojae | KJ175434 | EF203168 | EF202041 | MG517831 |
| CBS 100929 = NISL 1909 = IBT 21643 | Soy sauce, Japan | – | – | – | – | |
| CBS 100930 = NISL 1939 = IBT 21644 | Soy sauce, Japan | – | – | – | – | |
| CBS 100931 = NISL 1905 = IBT 21645 | Soy sauce, Japan | – | – | – | – | |
| CBS 100932 = IAM 2665 = IFO 4239 = NISL 1777 = IBT 21646 | Soy sauce, Japan | – | – | – | – | |
| CBS 100933 = NISL 1939 = IBT 21647 | Soy sauce, Japan | – | – | – | – | |
| CBS 100934 = IAM 2718 = IFO 4274 = RIB 1050 = NISL 1849 = IBT 21648 | Soy sauce, Japan | – | – | – | – | |
| CBS 100935 = NISL 1920 = IBT 21649 | Soy sauce, Japan | – | – | – | – | |
| CBS 100936 = IAM 2678 = RIB 1024 = IBT 21650 | Soy sauce, Japan (produces versicolorins) | – | – | – | – | |
| CBS 126.59 = IFO 5241 = IMI 191304 = Ohashi 1124 = IBT 3669 = IBT 3682 | Miso brewing, Okayama Agricultural Experiment Station, Japan | – | – | – | – | |
| CBS 133.52 = ATCC 9362 = CECT 2095 = IMI 087159 = NRRL 1947 = NRRL 1988 = NRRL 4841 = WB 4841 = IBT 3595 | Soy sauce, unknown origin | EF661546 | EF661482 | EF661517 | EF661450 | |
| DTO 173-C3 = IFM 46699 | Unknown source | – | MG517658 | MG518028 | MG517846 | |
| NRRL 5594 = IBT 4600 | Unknown source | – | – | – | – | |
| Aspergillus subflavus | CBS 143683T = DTO 326-E8 = S778 = CCF 4957 = NRRL 66254 = IBT 34939 | Soil, near Movile Cave, Romania, A. Nováková, 2013, ex type of Aspergillus subflavus | MH279429 | MG517773 | MG518143 | MG517964 |
| S843b | Moonmilk, Na Špičáku cave, Czech Republic, A. Nováková, 2013 | MH279449 | MG517792 | MG518164 | MG517983 | |
| Aspergillus tamarii | CBS 104.13T = NRRL 20818 = QM 9374 = IBT 3648 | Activated carbon, unknown origin, ex neotype of Aspergillus tamarii | AF004929 | EF661474 | EF661526 | EU021629 |
| CBS 133097 = DTO 213-H5 = NRRL 4959 | Unknown source, representative of Aspergillus tamarii var. crassus | MG662403 | MG517678 | MG518049 | MG517866 | |
| CBS 133393 = NRRL 4966 = IMI 016124 = IBT 3628 | Seed, cacao, unknown origin | EU021614 | EU021673 | EU021686 | EU021652 | |
| DTO 010-G9 = CBS 167.63 = NRRL 4680 = ATCC 15054 = IMI 172295 = QM 8903 = WB 4680 = IBT 22566 | Mouldy bread, India (ex type of Aspergillus indicus and A. terrricola var. indicus). Isolation of dihydrocanadensolide, fumaric acid, fumaryl-D,L-alanine, indazonic acid = cyclopiazonic acid, kojic acid, succinic acid and 3-nitropropionic acid show that these metabolites can be produced by A. tamarii (Birch et al., 1968) | MG662407 | MG517624 | MG518001 | MG517807 | |
| DTO 065-A4 | Indoor environment, Germany | MH279381 | MG517648 | MG517984 | MG517835 | |
| DTO 066-A1 | Corn kernels, Indonesia | - | MG517649 | MG517988 | MG517836 | |
| DTO 145-C3 | Indoor environment, Germany | MH279382 | MG517655 | MG517993 | MG517843 | |
| DTO 266-D7 | House dust, Mexico | - | MG517730 | MG518100 | MG517921 | |
| DTO 364-E3 | Air in chocolate factory, the Netherlands | MH279435 | MG517781 | MG518151 | MG517971 | |
| NRRL 425 | Unknown source, representative of Aspergillus lutescens nomen nudum | EF661558 | EF661475 | EF661524 | EU021648 | |
| NRRL 426 = DTO 010-H3 = CBS 579.65 = IBT 3681 = IBT 3826 = IBT 10827 | Unknown substrate, USA, ex neotype of Aspergillus terricola | EF661559 | EF661472 | EF661525 | EU021649 | |
| NRRL 4911 = CBS 484.65 = IBT 3659 | Air contaminant, Brazil, ex neotype of Aspergillus flavofurcatus | EF661565 | EF661473 | EF661527 | EU021651 | |
| Aspergillus togoensis | CBS 205.75T = LCP 67.3456 = NRRL 13551 = IBT 14899 = IBT 21943 | Decaying fruit of Landolphia sp., Central African Republic, ex type of Aspergillus togoensis | – | – | – | – |
| CBS 272.89 = DTO 034-C1 = NRRL 13550 = IBT 14989 = IBT 21943 | Seed, La Maboké, Central African Republic | AJ874113 | FJ491477 | FJ491489 | JN121479 | |
| Aspergillus transmontanensis | CBS 130015T = MUM 10.214 = IBT 32313 | Almond, Portugal, ex type of Aspergillus transmontanensis | JF412774 | HM803101 | HM803020 | HM802980 |
| MUM 10.205 | Almond, Portugal | JF412771 | HM803087 | HM803021 | HM802979 | |
| MUM 10.211 | Almond, Portugal | JF412772 | HM803102 | HM803023 | HM802968 | |
| MUM 10.221 | Almond, Portugal | JF446612 | HM803093 | HM803028 | HM802972 | |
| Aspergillus vandermerwei | CBS 612.78T = DTO 069-D2 = DTO 034-B5 = NRRL 5108 = IBT 13876 = CCF 5683 | Unknown source, Buenos Aires, Argentina, ex type of Aspergillus vandermerwei | EF661567 | EF661469 | EF661540 | MG517838 |
| DTO 199-A9 = CBS 129201 = DMSA 706 = IBT 16758 = CCF 5679 | Unknown source, USA, California | MH279390 | MG517661 | MG518032 | MG517849 | |
| DTO 210-F8 = CBS 132171 = IBT 16423 = RMF 7709 | Native shortgrass prairie, soil (1 m deep), Pawnee National Grassland, Colorado, USA | – | MG517675 | MG518046 | MG517863 | |
| DTO 363-F3 = NRRL 1237 = IBT 21072 = CCF 5602 | Unknown source | MH279434 | MG517780 | MG518150 | MG517970 | |
| DTO 368-B9 = IBT 16661 = CCF 5684 | Soil under crested wheat grass, 2 km south of Pryor, Colorado, USA | MH279436 | MG517783 | MG518153 | MG517973 | |
| DTO 368-C1 = NRRL 1236 = IBT 13865 = CCF 5685 | Unknown source | MH279437 | MG517784 | MG518154 | MG517974 | |
| DTO 368-C2 = CBS 126709 = RMF 9585 = IBT 20468 = CCF 5681 | Grassland, A1 soil horizon soil, Canyonlands National park, Utah, USA | MH279438 | MG517785 | MG518155 | MG517975 | |
| IBT 16662 | Soil under Senecio sp. (Asteraceae), Pablo Alto, Chaco Canyon, New Mexico, USA | MH279447 | MG517788 | MG518162 | MG517978 | |
| IBT 20491 | A1 soil horizon, Canyonlands National park, Utah, USA | MH279448 | MG517789 | MG518163 | MG517979 | |
Culture collections: ATCC; American Type Culture Collection, Mayland, USA, CBS, Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands, CCF: Culture Collection of Fungi, Prague, Czech republic, CCM: Czech Collection of Microorganisms, Brno, Czech Republic, CCTU: Culture Collection of Tabriz University, Iran, DSM: Deutsche Samlung von Mikroorganismen und Cell-kulturen, Braunschweig, Germany, DTO: The fungal working collection at Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands, IAM : Center for Cellular and Molecular Research, University of Tokyo, Tokyo, Japan (collection transferred to JCM), IFO (= NRBC) Institute of Fermentation, Osaka, Japan, IMI, CABI Fungal collection, Egham, UK, ITAL: Instituto de Technologia Alimentos, Campinas, Brazil, LCP: Laboratoire de Cryptogamie, Paris, France, KACC: Korean Agricultural Culture Collection, Seoul, South Korea, MUM: Micoteca da Univerdade do Minho, Portugal, NRRL, Northern Regional Research Lab, NCAUR, Peoria, Illinois, USA, QM: Quartermaster Collection, now at NRRL, Peoria, Illinois, USA, RIB: National Research Institute of Brewing, Higashihiroshima, Hiroshima, Japan, RMF: Rocky Mountain Fungi, collected by Martha Christensen, and situated in Laramie, Wyoming, USA, Thom: The original collection of Charles Thom, now at NRRL, WB: Wisconsin Bacteriology collection, Madison, Wisconsin, cultures now deposited at CBS, ATCC, IMI and IBT.
Results
Phylogenetic analysis
Various analyses were performed to study the phylogenetic relationship between species in section Flavi. Details on the number of included isolates, the length of the data sets and information on the used substitution model for each dataset are listed in Table 2.
Table 2.
Details of the length and substitution model of each data set.
| Description data set | No. isolates | BenA, length alignment | BenA, substitution model | CaM, length alignment | CaM, substitution model | RPB2, length alignment | RPB2, substitution model | Combined |
|---|---|---|---|---|---|---|---|---|
| Overview | 38 | 574 | K2 + G | 617 | TN93 + G | 880 | TN93 + G + I | 2071 |
| A. alliaceus-clade | 41 | 431 | K2 | 485 | TN93 + G | 760 | K2 + G | 1676 |
| A. flavus-clade | 133–138 | 481 | K2 | 529 | TN93 + G | 843 | TN93 + G | 1853 |
| A. leporis-clade | 16 | 510 | K2 + G | 555 | TN93 + G | 968 | TN93 + G | 2033 |
| A. nomius-clade | 25 | 504 | K2 + G | 528 | TN93 + G | 923 | TN93 | 1955 |
| A. tamarii-clade | 23 | 502 | K2 + G | 539 | HKY | 920 | K2 + G | 1961 |
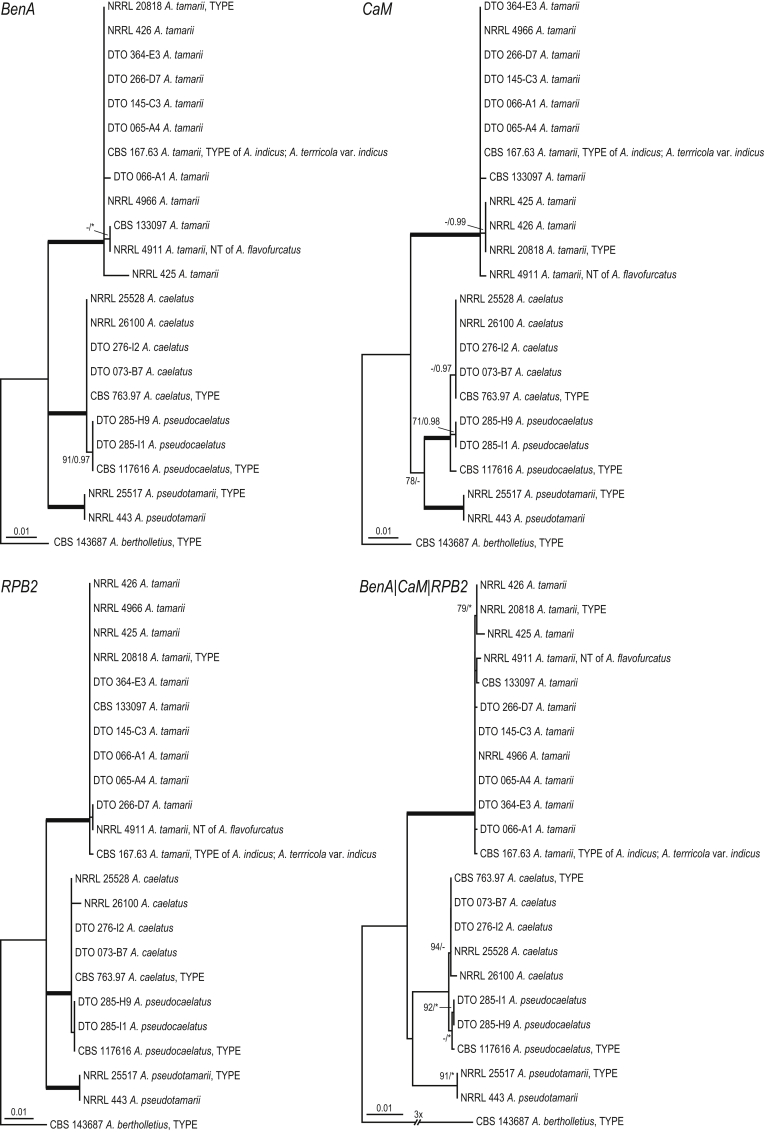
A phylogenetic study based on a combined data sets of loci (BenA, CaM, RPB2) was conducted to determine the relationship among Aspergillus section Flavi members. Aspergillus section Flavi could be subdivided into distinct eight clades: the A. alliaceus-, A. avenaceus-, A. bertholletius-, A. coremiiformis-, A. flavus-, A. leporis-, A. nomius- and A. tamarii-clade (Fig. 1). The A. flavus-clade is phylogenetically most closely related A. tamarii-clade and these clades form, together with the A. nomius- and the A. bertholletius-clades, a fully supported lineage. The phylogenetic relationship of the A. bertholletius-clade with the other clades remains partly unresolved in our analysis. In the ML analysis, the three A. bertholletius strains are placed with moderate statistical support (BS 83 %) in a basal position to the A. tamarii- and A. flavus-clades; however, no support was found in the Bayesian analysis (< 0.95 pp). The A. alliaceus- and A. coremiiformis-clades are also phylogenetically related (92 % BS, 1.00 pp) and these clades form a sister lineage to the A. flavus-, A. tamarii-, A. bertholletius- and A. nomius-clades. Aspergillus leporis and related species (A. leporis-clade) take a basal position to aforementioned clades and the A. avenaceus-clade, only represented by A. avenaceus, is basal in section Flavi.
Fig. 1.
Phylogeny inferred from a concatenated nucleotide data set (partial BenA, CaM and RPB2 sequences) using ML analysis showing the relationship of species accommodated in Aspergillus section Flavi. The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
The A. flavus-clade is the most species-rich clade of section Flavi and contains 15 species (Fig. 2, Fig. 3), including the five new species described in this manuscript (see Taxonomy; A. aflatoxiformans, A. austwickii, A. cerealis, A. pipericola, A. subflavus). Analysis of the combined data set reveals four well-supported lineages in the A. flavus-clade. One main lineage is centered on A. flavus and contains A. aflatoxiformans, A. austwickii, A. cerealis, A. flavus, A. minisclerotigenes, A. oryzae and A. pipericola and the other main lineage (centered on A. parasiticus) includes A. arachidicola, A. novoparasiticus, A. parasiticus, A. sergii, A. sojae and A. transmontanensis. Aspergillus mottae has a basal position to the A. flavus and A. parasiticus lineages and A. subflavus is basal to all A. flavus-clade species. Almost all species could be resolved in the phylogenetic analysis of the combined data set. There are two exceptions: A. oryzae resides in a clade with A. flavus and A. sojae forms a clade with A. parasiticus. With the exception of A. flavus/A. oryzae and A. parasiticus/A. sojae, almost all species could be recognised using BenA, CaM or RPB2 sequences only. The exception is A. novoparasiticus and this species shares BenA sequences with A. parasiticus isolates. Strains CBS 485.65 (ex-type of A. flavus var. columnaris and A. flavus var. asper), CBS 501.65 (A. subolivaceus), CBS 542.69 (A. kambarensis), CBS 120.51 (A. thomii) and CBS 110.55 (A. fasciculatus) belong to the A. flavus/A. oryzae clade and CBS 260.67 (A. parasiticus var. globosus), CBS 580.645 (A. terricola var. americana) and CBS 822.72 (A. toxicarius) reside in the A. parasiticus/A. sojae clade. Two interesting A. flavus strains (from air, Korea) that produce aflatoxins of the B and G type (CBS 143688, CBS 143689) cluster in all analyses with other A. flavus/A. oryzae strains. Also two strains with small sized sclerotia (DTO 281-H8; NRRL 3251) belong to the A. flavus/A. oryzae lineage.
Fig. 2.
ML Phylogeny showing the relationship of species accommodated in the A. flavus-clade (left, BenA; right, CaM). The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
Fig. 3.
Phylogeny showing the relationship of species accommodated in the A. flavus-clade (left, RPB2; right, combined data set of BenA, CaM and RPB2). The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
Phylogenetic analysis reveals the presence of four species (A. caelatus, A. pseudocaelatus, A. pseudotamarii and A. tamarii) in the A. tamarii-clade (Fig. 4). The genetic distance between A. caelatus (CBS 763.97, DTO 073-B7, DTO 276-I2, NRRL 25528, NRRL 26100) and A. pseudocaelatus (DTO 285-H9, DTO 285-I1, CBS 117616) strains is low. In the phylogeny based on the combined data set, these strains resolve in two distinct clades. This clade is fully supported in the Bayesian analysis (1.00 pp); however, it lacks confident bootstrap support in the ML analysis (< 70 %). Representative strains of A. flavofurcatus (NRRL 4911), A. indicus and A. terricola var. indicus (CBS 167.63) cluster together with the type of A. tamarii (NRRL 20818) in all analyses.
Fig. 4.
Phylogeny showing the relationship of species accommodated in the A. tamarii-clade. The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
Three species are accommodated in the A. nomius-clade: A. luteovirescens, A. nomius and A. pseudonomius (Fig. 5). Three well-supported, distinct clades could be recognised in the BenA analysis, representing the species accommodated in this clade. Not all species were resolved in the CaM and RPB2 analysis. The statistical support in the CaM phylogram was low. Aspergillus nomius strain DTO 321-F2 clustered with the included A. pseudonomius strains; however, statistical support was lacking. Phylogenetic analysis of the RPB2 data set could not resolve A. nomius and A. pseudonomius and strains of those species appear intermixed on one well-supported branch. The ex-type of A. zhaoqingensis (CBS 399.93) clusters together with the A. nomius strains in three out of the four analyses (BenA, CaM and combined analysis), and the ex-type of A. bombycis NRRL 26010T clusters with A. luteovirescens strains (incl. CBS 620.95NT).
Fig. 5.
Phylogeny showing the relationship of species accommodated in the A. nomius-clade. The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
Four distinct groups in the A. alliaceus-clade can be recognised after assessment of the phylograms using the Genealogical Concordance Phylogenetic Species Recognition (GCPSR; Taylor et al. 2000) concept (Fig 6, Fig. 7). These groups represent two known (A. alliaceus, A. lanosus) and two new species (described here as A. neoalliaceus and A. vandermerwei). The deeper nodes in the phylograms often have a low statistical support and the relationship among species in the A. alliaceus-clade therefore remains unknown. A high BenA, CaM and RPB2 sequence diversity is present in the A. vandermerwei. Following the GCPSR concept, two groups can be recognised in A. vandermerwei: one includes CBS 129201, CBS 132171 and DTO 368-B9, and the other contains CBS 126709, DTO 368-C1, DTO 363-F3, IBT 20491, IBT 16662 and NRRL 5108T. The ex-type strain of A. albertensis, NRRL 20602, resides in the clade containing A. alliaceus isolates.
Fig 6.
ML Phylogeny showing the relationship of species accommodated in the A. alliaceus-clade (left, BenA; right, CaM). The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
Fig. 7.
Phylogeny showing the relationship of species accommodated in the A. alliaceus-clade (left, RPB2; right, combined data set of BenA, CaM and RPB2). The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
A set of strains isolated from soil of Aspear Island in Urmia Lake (Iran) formed a distinct lineage related to A. leporis and the name A. aspearensis is proposed for this group of isolates (Fig. 8). The third species in this clade is the recently described species A. hancockii (Pitt et al. 2017). All species can be recognised using the GCPSR concept.
Fig. 8.
Phylogeny showing the relationship of species accommodated in the A. leporis-clade. The bar indicates the number of substitutions per site. The BI posterior probabilities values and bootstrap percentages of the ML analysis are presented at the node (BS/pp). Values less than 70 % bootstrap support in the ML analysis and less than 0.95 posterior probability in the Bayesian analysis are indicated with a hyphen. Branches with high support (> 95 % bs; 1.00 pp) are thickened and the BS and pp values indicated with an asterisks.
Extrolite analysis
An overview of mycotoxins and other extrolites produced by Aspergillus section Flavi is given in Table 3, Table 4. The A. avenaceus- and A. leporis-clades are basal to the other clades in section Flavi (Fig. 1), but do not have the ability to produce aflatoxins or ochratoxins. Furthermore, A. avenaceus does not produce kojic acid, an extrolite produced by the majority of species in section Flavi (Table 3). Aflatoxins or precursors of aflatoxins are produced in all the other clades (the A. flavus-, A. tamarii-, A. bertholletius-, A. nomius-, A. alliaceus- and A. coremiiformis clades). Ochratoxin A and B are only found in the A. alliaceus-clade. Among the species in Aspergillus section Flavi, two species produced aflatoxin B1 and B2 only: A. pseudotamarii and A. togoensis. Sixteen species produced aflatoxin B1, B2, G1 and G2: A. aflatoxiformans, A. arachidicola, A. austwickii, A. cerealis, A. luteovirescens, A. minisclerotigenes, A. mottae, A. nomius, A. novoparasiticus, A. parasiticus, A. pipericola, A. pseudocaelatus, A. pseudonomius, A. sergii, A. transmontanensis and some strains of A. flavus (Table 3, Supplementary Fig. S1). One strain of A. bertholletius produced the aflatoxin B1 precursor O-methylsterigmatocystin (Taniwaki et al. 2012, and this result was confirmed here). Seven strains of A. flavus sensu stricto from Korea were found to produce aflatoxins of the B and G type (Table 3). Most isolates of A. alliaceus, A. neoalliaceus and A. vandermerwei produced large amounts of ochratoxin A (Table 3; Supplementary Table S1, Supplementary Fig. S2). One strain of A. sojae and two strains of A. alliaceus produced versicolorins (Table 3), precursors of the aflatoxins. Another important mycotoxin, tenuazonic acid, is produced by eight species (A. bertholletius, A. caelatus, A. luteovirescens, A. nomius, A. pseudocaelatus, A. pseudonomius, A. pseudotamarii and A. tamarii) (Supplementary Fig. S3). The related mycotoxin cyclopiazonic acid was produced by 14 species: A. aflatoxiformans, A. austwickii, A. bertholletius, A. cerealis, A. flavus, A. hancockii (only speradine F found in this species), A. minisclerotigenes, A. mottae, A. oryzae, A. pipericola, A. pseudocaelatus, A. pseudotamarii, A. sergii and A. tamarii (Table 4, Supplementary Fig. S4).
Table 3.
Mycotoxin and other extrolite production by Aspergillus section Flavi species.
| Species | Extrolites reported in literature | Extrolites detected in this study | Examined strains |
|---|---|---|---|
| Aspergillus aflatoxiformans | – | Aflatoxin B1, B2, G1, G2, aflatrems, aflavarins, aflavinines, aspergillic acid, aspirochlorin, cyclopiazonic acid, kojic acid, paspaline, paspalinine, versicolorins, metabolite gfn (UV absorbtions 240 nm & 397 nm, RI 1148) | DTO 228-G1, DTO 228-G2T DTO 228-G3, DTO 228-G4 DTO 228-G5, DTO 228-G6, DTO 228-G7, DTO 228-H2, DTO 228-H3, DTO 228-H6, DTO 228-H7, CBS 133923, CBS 133924, CBS 133264, CBS 133265, CBS 133925, DTO 087-A2, CBS 121.62, DTO 010-H7 |
| A. alliaceus | Anominine (Laakso et al., 1994, Nozawa et al., 1994), asperlicin A-E (Liesch et al., 1985, Liesch et al., 1988), 7-O-demethyl-3,8′-bisiderin, 7-O-demethyl-6,6′-bisiderin (Nozawa et al. 1994), 14-(N,N-dimethyl-L-leucinoxy)paspalinine, 14-hydroxypaspalinine (Junker et al. 2006), isokotanins A-C (Laakso et al. 1994), kojic acid (Manabe et al. 1984), kotanin (Nozawa et al. 1994), ochratoxin A and B (Ciegler, 1972, Bayman et al., 2002), paspaline (Laakso et al. 1994) | Anominine (8/13 strains), antarone A (4/13 strains), asperlicins (5/13 strains), isokotanins (7/13 strains), kojic acid (13/13 strains), met I1 (10/13 strains), ochratoxin A & B (13/13 strains), paspaline (10/13 strains), versicolorin (2/13 strains: DTO 363-F1, DTO 363-E8). For more details, see Supplementary Table S1. | CBS 542.65T, CBS 511.69, DTO 326-D5, DTO 363-E8, DTO 363-E9, DTO 363-F1, DTO 363-F2, DTO 368-C4, IBT 21770, NRRL 315, NRRL 316, NRRL 317, NRRL 20602 |
| A. arachidicola | Aflatoxin B1, B2, G1 & G2 (Pildain et al. 2008, aspergillic acid (Pildain et al. 2008), chrysogine (Pildain et al. 2008), ditryptophenaline (Varga et al. 2011), kojic acid (Pildain et al. 2008), parasiticolides (Pildain et al. 2008) | Aflatoxin B1, B2, G1, G2, aspergillic acid (only in CBS 117613 & CBS 117614), chrysogine (chrysogine precursor in CBS 117614), ditryptophenaline, kojic acid, miyakamides, parasiticolides | CBS 117610, CBS 117611, CBS 117612, CBS 117613, CBS 117614, CBS 117615 |
| A. aspearensis | – | An aflavinine, kojic acid, mevinolins, paspalinines | DTO 203-D9, DTO 203-E1, DTO 203-D4 |
| A. austwickii | – | Aflatoxin B1, B2, G1, G2, aflatrems, aflavarins, cyclopiazonic acid, kojic acid, paspaline, paspalinine, versicolorins, metabolite gfn | DTO 228-F7T, DTO 228-F8, DTO 228-F9, DTO 228-G8 |
| A. avenaceus | Avenaciolide (Brookes et al. 1963), aspirochlorine (Varga et al. 2011), 4-isoavenaciolide (Turner, 1971, Turner and Aldridge, 1983), 3-nitropropionic acid (Brookes et al. 1963) | An altersolanol (only in IMI 238253 = IBT 19369 & IMI 232294 = IBT 19371), aspirochlorin, avenaciolides, pseurotin A (only in IMI 093340 = IBT 19372), 2-(4-hydroxyphenyl)-2-oxo acetaldehyde oxime (only in NRRL 4517 = IBT 18842) | CBS 109.46T, IMI 093340, IMI 232294, IMI 238253, NRRL 4517 |
| A. bertholletius | Cyclopiazonic acid, kojic acid, O-methylsterigmatocystin, parasiticolide, tenuazonic acid, ustilaginoidin C (Taniwaki et al. 2009) | Cyclopiazonic acid, kojic acid, O-methylsterigmatocystin (only the ex-type strain), parasiticolides (only two strains: IBT 31546, IBT 31739), tenuazonic acid, ustilaginoidin C | IBT 29228, IBT 30618, IBT 30617, IBT 30619, IBT 29227 |
| A. caelatus | Aspirochlorin (Pildain et al. 2008), kojic acid (Frisvad & Samson 2000), tenuazonic acid (Varga et al. 2011) | An altersolanol, aspirochlorin, kojic acid, tenuazonic acid, in addition to an indole alkaloid (“alkca”)(RI 928) that has only been found in A. caelatus, A. pseudocaelatus and A. pseudotamarii | CBS 763.97T, CBS 764.97, NRRL 25566, NRRL 25567, NRRL 25568, NRRL 25569 |
| A. cerealis | – | Aflatoxin B1, B2, G1, G2, aflatrems, aflavarins, aflavazole, cyclopiazonic acid, kojic acid, paspaline, paspalinine, versicolorins | CBS 143674T, DTO 228-E6, DTO 228-E8, DTO 228-E9, DTO 228-F1, DTO 228-F2, DTO 228-F3, DTO 228-F4, DTO 228-F5, DTO 228-F6 |
| A. coremiiformis | Indole alkaloids (Varga et al. 2011) | No known extrolites found | CBS 553.77T |
| A. flavus | Aflatoxin B1 and B2 (Nesbitt et al., 1962, Codner et al., 1963, Varga et al., 2009, Rank et al., 2012 and many others), aflatrem & β-aflatrem (Gallagher and Wilson, 1978, TePaske et al., 1992, Rank et al., 2012, Sun et al., 2014), aflavarin A-C (TePaske et al. 1992), aflavazole (TePaske et al. 1990), asparasones (Cary et al., 2014, Malysheva et al., 2014, Chang et al., 2017), aspergillic acid (White and Hill, 1943, Assante et al., 1981), aspergillomarasmine A & B (Robert et al., 1962, Haenni et al., 1965), aspirochlorins (Sakata et al., 1982, Sakata et al., 1987, Klausmeyer et al., 2005, Rank et al., 2012), bright-greenish-yellow-fluorescence (6,6′-bis[5-hydroxy-2-(hydroxymethyl)-4H-pyran-4-one], “dikojic acid”) (Zeringue et al. 1999), cAATrp (Uka et al. 2017), cyclopiamide A-G & J (Ma et al., 2015, Uka et al., 2017), α-cyclopiazonic acid, β-cyclopiazonic acid, iso-α-cyclopiazonic acid, α-cyclopiazonic acid imine (Luk et al., 1977, Rank et al., 2012, Sun et al., 2014, Uka et al., 2017), (S)-(-)-6,8-di-O-methylcitreoisocoumarin (Sun et al. 2014), ditryptophenaline (Springer et al., 1977, Rank et al., 2012, Sun et al., 2014), gliotoxin (Lewis et al., 2005, Kupfahl et al., 2008), 3-hydroxy-speradine A (Uka et al. 2017), kojic acid (Birkinshaw et al., 1931, Manabe et al., 1984, Rank et al., 2012, Sun et al., 2014), leporins (Sun et al., 2014, Arroya-Manzanares et al., 2015), miyakamides (Shiomi et al. 2002), 3-nitropropionic acid (Bush et al., 1951, Becker and Schmidt, 1964, Doxtater and Alexander, 1966, Konoshita et al. 1968, Hatcher and Schmidt, 1971, Iwasaki and Kosikowski, 1973), 2-oxo-cyclopiazonic acid (Uka et al. 2017), parasiticolide A (Shiomi et al. 2002), paspaline, β-PC-M6, 13-desoxypaxilline, 4b-deoxy-β-aflatrem, 9-isopentenylpaxilline D, paspalicine & paspalinine (Cole et al., 1981, Rank et al., 2012, Sun et al., 2014), penicillin G (Bush and Goth, 1943, Bush et al., 1945, Adler and Wintersteiner, 1948, Guida, 1948, Blinc and Johanides, 1956), speradine A-D, F & H (Ma et al., 2015, Uka et al., 2017), ustiloxin (Umemura et al., 2013a, Umemura et al., 2013b, Umemura et al., 2014, Ye et al., 2016). Reported to be produced by Aspergillus flavus CBS 131.61: Aflatoxin B1, G1, aspergillic acid, aspyrone, betaine, chrysogine, diacetyl parasiticolide A, flufuran, gregatin B, hydroxysydonic acid, nicotinic acid, phomaligin A, spinulosin and terrein (Saldan et al. 2018) | Aflatoxin B1 and B2, aflatrems (only in sclerotium producers), aflavarins (only in sclerotium producers), aflavinines (only in sclerotium producers), asperfuran, aspergillic acid, aspirochlorin, citreoisocoumarin, cyclopiazonic acids, ditryptophenaline, flavimin, kojic acid, miyakamides, paspaline & paspalinine (only in sclerotium producers), ustilaginoidin C (ATCC 26850, CBS 116.48, CBS 113.49, CBS 120.51, CBS 110.55, CBS 131.62, CBS 117.62, CBS 118.62, CBS 119.62, CBS 242.65, CBS 501.65, CBS 569.65T = CBS 100927 = NRRL 1957, CBS 625.66, CBS 542.69, CBS 289.95, CBS 816.96, CBS 970.97, CBS 117625, CBS 117632, CBS 127422, NRRL 453, NRRL 3251, NRRL 3357, NRRL 5565, NRRL 6551, NRRL 6556, NRRL 29254). Some columnar isolates of A. flavus produce aflatoxin B2 (IBT 12654, NRRL 5821). Special Korean strains: DTO 359-E4: Aflatoxin B1, B2, G1, G2, kojic acid, ustilaginoidin C; DTO 359-D7, DTO 359-D8, DTO 359-D9, DTO 359-E1, DTO 359-E2, DTO 359-E8: aflatoxin B1, B2, G1, G2, cyclopiazonic acid, flavimin2, kojic acid, ustilaginoidin C | ATCC 26850, CBS 116.48, CBS 113.49, CBS 120.51, CBS 110.55, CBS 131.62, CBS 117.62, CBS 118.62, CBS 119.62, CBS 242.65, CBS 501.65, CBS 569.65T = CBS 100927 = NRRL 1957, CBS 625.66, CBS 542.69, CBS 289.95, CBS 816.96, CBS 970.97, CBS 117625, CBS 117632, CBS 127422, NRRL 453, NRRL 3251, NRRL 3357, NRRL 5565, NRRL 6551, NRRL 6556, NRRL 29254 |
| A. hancockii | Dehydroterrestric acid, eupenifeldin, fumitremorgin A, hancockiamide A-F, 7-hydroxytrichothecolone, kojic acid, onychocin A & B, speradine F (Pitt et al. 2017) | An aflavarin, dehydroterrestric acid, fumitremorgin A, hancockiamide A, 7-hydroxytrichothecolone, onychocin A & B, a speradine | CBS 142002, CBS 142001 |
| A. lanosus | Griseofulvin (Frisvad & Samson, 2000), kojic acid (Frisvad & Samson, 2000), ochratoxin A & B (Baker et al., 2003, Palumbo et al., 2007) | An altersolanol, an asperlicin, griseofulvin, kojic acid, met I1. For more details, see Supplementary Table S1. | CBS 650.74T |
| A. leporis | Antibiotic Y (Frisvad & Samson, 2000), kojic acid (Frisvad & Samson, 2000), leporins (TePaske et al. 1991), leporizines (Reategui et al. 2013), pseurotin A (Frisvad & Samson, 2000) | Antibiotic Y, clavatols, 7-hydroxytrichothecolone?, kojic acid, leporine A, leporiziznes, paspalines, pseurotin A | CBS 151.66, CBS 349.81, ATCC 76617, IBT 16309, IBT 16585 |
| A. luteovirescens | Aflatoxin B1, B2, G1, G2 (Pildain et al. 2008), aspergillic acid (Varga et al. 2011), kojic acid (Morton et al., 1945, Varga et al., 2011) | Aflatoxin B1, B2, G1, G2, an altersolanol (only in NRRL 29235 & NRRL 29253), aspergillic acid, chrysogine (only in NRRL 29253), kojic acid, sporogen AO1, tenuazonic acid (in IBT 31534, NRRL 29235, NRRL 29237). CBS 620.95T only produced kojic acid. | CBS 620.92T, CBS 117187, DTO 073-C2, DTO 073-C3, DTO 073-C5, IBT 31534, NRRL 25593, NRRL 29237, NRRL 29253 |
| A. minisclerotigenes | Aflatoxin B1, B2, G1, G2, aflavarins, aflatrems, aflavinines, aspergillic acid, cyclopiazonic acid, kojic acid, paspalinine (Pildain et al. 2008) | Aflatoxin B1, B2, G1, G2, aflatrems, aflavarins, aflavazole (in DTO 228-H1 & IBT 27213), aflavinines, aspergillic acid, cyclopiazonic acid, kojic acid, parasiticolides, paspalinine | CBS 117635T, CBS 117620, CBS 117634, CBS 117637, CBS 117639, DTO 228-G9, DTO 228-H1, DTO 228-H5, IBT 27213, NRRL 6444 |
| A. mottae | Aflatoxin B1, B2, G1, G2 (Soares et al. 2012) | Aflatoxin B1, B2, G1, G2, an aflavinin, aspergillic acid, cyclopiazonic acid, kojic acid, 3-O-methylsterigmatocystin, parasiticol, paspalinine, versicolorins | CBS 130016T = DTO 223-C8 = IBT 32309 = MUM 10.231 |
| A. neoalliaceus | – | Anominine (6/9 strains), brefeldin A (5/9 strains), kojic acid (9/9 strains), ochratoxin A and B (9/9 strains), paspaline (7/9 strains). For more details, see Supplementary Table S1. | CBS 143681T, CBS 134375, DTO 326-D7, DTO 326-D8, DTO 326-D1, DTO 326-E4, DTO 326-E2, DTO 326-E7, DTO 326-D6, DTO 326-E5 |
| A. nomius | Aflatoxin B1, B2, G1, G2 (Kurtzman et al. 1987), anominine (Gloer et al., 1989, Bradshaw et al., 2010), aspergillic acid (Frisvad & Samson 2000), aspernomine (Staub et al. 1992), kojic acid (Frisvad & Samson 2000), paspaline (Staub et al. 1992), pseurotin (Frisvad & Samson 2000), tenuazonic acid (Frisvad & Samson 2000) | Aflatoxin B1, B2, G1, G2, anominine, aspergillic acid, aspernomine, kojic acid, a miyakamide, 3-O-methylsterigmatocystin, parasiticol, paspaline, paspalinine, pseurotin A, tenuazonic acid, versicolorins and other aflatoxin precursors | CBS 260.88T, CBS 399.93, CBS 117629, IMI 190557, NRRL 13138, NRRL 3161 |
| A. novoparasiticus | Aflatoxins B1, B2, G1, G2 (Gonçalves et al., 2012a, Gonçalves et al., 2012b) | Aflatoxins B1, B2, G1, G2, (aspirochlorin, ditryptophenaline, kojic acid, miyakamides, parasiticolide, and a tetracyclic compound | CBS 126849, CBS 126830 |
| A. oryzae | Aflavinines (Rank et al. 2012), asperfuran (Pfefferle et al. 1990), aspergillomarasmins (Robert et al., 1962, Barbier et al., 1963), asperopterin A & B (Matsuura et al. 1972), aspirochlorins (Sakata et al., 1983, Champhamjon et al., 2014), cyclopiazonic acid and speradines (Orth, 1977, Tokuoka et al., 2015), 14-deacetyl parasiticolide A & B and dideacetyl parasiticolide A, confertifolin, dideacetyl astellolide A & B (Rank et al., 2012, Shinohara et al., 2016a, Shinohara et al., 2016b), 13-desoxypaxilline (Rank et al. 2012), ditryptoleucine (Rank et al. 2012), kojic acid (Birkinshaw et al. 1931), kojistatin (Sato et al., 1996, Yamada et al., 1998), maltoryzin (Iizuka and Iida, 1962), 3-nitropropionic acid (Nakamura and Shimoda, 1954, Yokotsuka et al., 1969, Orth, 1977), oryzamides (Rank et al. 2012), paspaline and β-PC-M6 (Rank et al. 2012), penicillin (Saito 1946–47), speradine B-F (Hu et al., 2014a, Hu et al., 2014b), sporogen AO1 (Tamogami et al. 1996), TMC-2A, B & C (Nonoka et al., 1977, Asai et al., 1998) | Asperfuran (CBS 102.22, CBS 134.52, IBT 3629), aspirochlorin (CBS 102.07T, CBS 134.52, CBS 570.65, CBS 819.72, RIB 40), citreoisocoumarin (CBS 102.22, CBS 570.65, CBS 205.89, NRRL 6270), a cyclopiamide (IBT 3593, IBT 3629, NRRL 695), cyclopiazonic acid (CBS 102.07T = CBS 110.47T, CBS 570.65, CBS 205.89, IBT 3593, IBT 3629, NRRL 484), ditryptoleucine (RIB 40), kojic acid (CBS 102.07, CBS 134.52, CBS 570.65, CBS 205.89, IBT 3595, IBT 3629, NRRL 695), miyakamides / oryzamides (CBS 102.07T = CBS 110.47T, CBS 570.65, RIB 40), parasiticolides / astellolides (CBS 570.65, CBS 819.72, CBS 205.89, NRRL 695, RIB 40), paspalines (RIB 40), sporogen AO1 (NRRL 6270). According to verified strains of A. oryzae, isolates of the species can also produce penicillins and 3-nitropropionic acid | CBS 102.07T (= CBS 110.47T = CBS 100925T), CBS 102.22, CBS 134.52, CBS 570.65, CBS 205.89, IBT 3593, IBT 3629, NRRL 484, NRRL 695, NRRL 6270, RIB 40 |
| A. parasiticus | Aflatoxin B1, B2, G1, G2 (Codner et al., 1963, Schroeder, 1966, Basaran and Demirbas, 2010), asparasone A (Sobolev et al. 1997), aspergillic acid (Assante et al. 1981), aspersitin (Hamasaki et al. 1975), dibutylphthalate (an artefact?) (Basaran & Demirbas 2010), fumagillol (Basaran & Demirbas 2010), italicic acid (Basaran & Demirbas 2010), kojic acid (Birkinshaw et al., 1931, Basaran and Demirbas, 2010), parasperone and ustilaginoidin C (Brown et al. 2003), parasitenone (Son et al. 2002), parasiticol (Stubblefield et al. 1970), parasiticolide A (= astellolide A) (Büchi et al., 1983, Rank et al., 2012), penicillin G (Arnstein & Cook 1947), pyrogallol (Basaran & Demirbas 2010), sequioatones (Stierle et al., 1999, Stierle et al., 2001), sequoiamonascins (Stierle et al. 2003), sorbicillin (Basaran & Demirbas 2010) | Aflatoxin B1, B2, G1, G2, aspergillic acid, kojic acid, parasperone, parasiticol, parasiticolide A and B | CBS 100926T, CBS 822.72, CBS 580.65, CBS 260.67, CBS 921.70, NRRL 6433, NRRL 13005 |
| A. pipericola | – | Aflatoxin B1, B2, G1, G2, aflatrem, aflavinins, aflavarins, cyclopiazonic acid, paspaline, paspalinine | CBS 143680T |
| A. pseudocaelatus | Aflatoxin B1, B2, G1, G2, cyclopiazonic acid, kojic acid (Varga et al. 2011) | Aflatoxin B1, B2, G1, G2, aspirochlorin, cyclopiazonic acid, ditryptophenaline, kojic acid, tenuazonic acid, “alkca” | CBS 117616, IBT 29230, DTO 350-B8 |
| A. pseudonomius | Aflatoxin B1, chrysogine, kojic acid (Varga et al. 2011) | Aflatoxin B1, B2, G1, G2 (ex type isolate only produce type B aflatoxins), aspergillic acid, chrysogine, kojic acid, a miyakamide, tenuazonic acid | CBS 119388T, DTO 079-I4, IBT 12657, IBT 32759, NRRL 5919 (= IBT 23354), NRRL 6343 = IBT 4496 = IBT 4985 |
| A. pseudotamarii | Aflatoxin B1, B2, cyclopiazonic acid, kojic acid (Ito et al., 2001, Varga et al., 2011) | Aflatoxin B1, B2, aflavinines, an altersolanol (in CBS 766.97 CBS 117625 & CBS 117628), aspirochlorin (in CBS 766.97 & IBT 30530), cyclopiazonic acid, kojic acid, paspaline & paspalinine (in CBS 117628), tenuazonic acid, “alkca” | CBS 766.97, CBS 117625, CBS 117628; IBT 30530, IBT 30531 |
| A. sergii | Aflatoxin B1, B2, G1, G2 (Soares et al. 2012) | Aflatoxin B1, B2, G1, G2, aflatrem, aflavazole, an aflavarin, aflavinins, asperfuran, aspergillic acid, cyclopiazonic acid, kojic acid, paspalinine, versicolorins | CBS 130017T, DTO 223-C9 |
| A. sojae | Asperfuran (Varga et al. 2011), aspergillic acid (Pildain et al. 2008), aspirochlorin, chrysogine (Varga et al. 2011), kojic acid (Tanaka et al. 2002) | Asperfuran, aspergillic acid, aspirochlorin, chrysogine, kojic acid, miyakamides, versicolorins (only CBS 100936) | CBS 100928T, CBS 133.52, CBS 126.59, CBS 100929, CBS 100930, CBS 100932, CBS 100933, CBS 100934, CBS 100935, CBS 100936, NRRL 5594 |
| A. subflavus | – | Aflavinines, aspirochlorin, kojic acid, a parasiticolide | CBS 143683T |
| A. tamarii | Aspirochlorin (Berg et al. 1976), (-)-canadensolide (Berg et al. 1976), cyclopiazonic acid (Dorner 1983), dihydrocanadensolide, fumaric acid, fumaryl-D,L-alanine (Birch et al. 1968), fumigaclavine A (Jahardhanan et al. 1984), kojic acid (Birkinshaw et al., 1931, Manabe et al., 1984), 3-nitropropionic acid (Birch et al. 1968), speradine A (Tsuda et al. 2003), succinic acid (Birch et al. 1968) | Aspirochlorin (8/15 strains), citreoisocoumarin (2/15 strains), cyclopiazonic acid (9/15 strains), kojic acid (13/15 strains), tenuazonic acid (4/15 strains) | CBS 103.14T, CBS 104.14, CBS 129.49, CBS 109.63, CBS 167.63, CBS 484.65, CBS 575.65, CBS 579.65, CBS 591.68, CBS 117626, CBS 126844, IBT 29248, IBT 29229, NRRL 4860, NRRL 8101 |
| A. togoensis | Aflatoxin B1 (Rank et al. 2011), sterigmatocystin (Wicklow et al. 1989) | Aflatoxin B1, a bisiderin, paspaline, paspalinine, sterigmatocystin (CBS 205.75T), paxilline (CBS 272.89) | CBS 205.75, CBS 272.89 |
| A. transmontanensis | Aflatoxin B1, B2, G1, G2 (Soares et al. 2012) | Aflatoxin B1, B2, G1, G2, aspirochlorin, kojic acid, a miyakamide | CBS 130015T |
| A. vandermerwei | – | An altersolanol, anominine, an asperlicin, aspirochlorin, bostrycin?, brefeldin A, kojic acid, isokotanins, ochratoxin A, ochratoxin B. Griseofulvin produced by CBS 126708. For more details, see Supplementary Table S1. | DTO 069-D2T, IBT 16662, IBT 20491, CBS 612.78, CBS 129201, DTO 363-F3, DTO 368-B9, CBS 126709, DTO 368-C1, CBS 132171 |
Table 4.
Mycotoxin producing species in Aspergillus section Flavi.
Morphology and physiology
Species in section Flavi produce spreading, transparent colonies on CREA that measure (25–)35–50(–55) mm after 7 d and acid production is generally absent. Weak acid production is present in some strains of certain species (A. caelatus, A. pseudocaelatus, A. pseudotamarii, A. tamarii); however, this is not a consistent character at species level. A colony diameter larger than 5 mm after 7 d incubation on CYA at 42 °C (CYA42°C) was observed in A. aflatoxiformans, A. arachidicola, A. austwickii, A. cerealis, A. flavus, A. minisclerotigenes, A. novoparasiticus, A. oryzae, A. parasiticus, A. sergii and A. sojae (Fig. 9, Fig. 10, Fig. 11, Fig. 12, Table 5). Some strains inconsistently grew on CYA42°C: certain strains of A. alliaceus, A. lanosus, A. neoalliaceus, A. nomius, A. pipericola produced restricted colonies on CYA42°C (1–5 (–8) mm), while no growth was observed in other isolates. Aspergillus coremiiformis and A. togoensis did not grown on CYA incubated at 37 °C and A. avenaceus produced restricted colonies at that temperature (7 mm after 7 d); all other species grow well on CYA37 °C. Species belonging to section Flavi grow rapidly and generally attain a diameter of more than 50 mm on CYA, MEA and YES after 7 d; the exception is A. coremiiformis (CYA 30 mm, MEA 46 mm, YES 48 mm). The conidial colour can be in shades of brown, green and yellow. The majority of species have conidia in shades of (dark) yellow-green (e.g. A. flavus, A. austwickii, A. arachidicola, A. nomius, A. parasiticus, A. transmontanensis); conidia in shades of brown are produced by e.g. A. bertholletius, A. caelatus, A. pseudocaelatus, A. tamarii and yellow shades are present in isolates of A. alliaceus, A. lanosus, A. neoalliaceus and A. vandermerwei. A majority of species, 28 out of 33 species in Aspergillus section Flavi, can produce sclerotia (Table 5); however, not always on the media used in this study. Sclerotium production was often best on CYA incubated at 25 °C or 37 °C, followed by MEA and YES. The sclerotia produced by section Flavi species become dark brown or black coloured at age and have different shapes and sizes. Examples are shown in Fig. 13, Fig. 14. Species belonging to the A. flavus-clade generally produce globose to ellipsoidal sclerotia that can be large (e.g. A. flavus, A. parasiticus, A. transmontanensis, 400–700 (–1000) μm; A. subflavus 375–650 μm), intermediate (A. sergii 300–550 μm) or small (A. aflatoxiformans, A. austwickii, A. cerealis, A. minisclerotigenes, A. pipericola 100–375 μm; A. mottae 150–375 μm). Although the majority of A. flavus strains produce large-sized sclerotia, some isolates have sclerotia less than 350 μm in diam (Fig. 13, Table 5). Species in the A. alliaceus-clade produce large sclerotia (1000–2500 × 500–1200 μm) that are oblong to oval shaped, brownish black coloured, which occasionally have a white tip on the top (Fig. 14A). The sclerotia produced by species in the A. leporis-clade are ellipsoidal or irregular shaped and vary in size. Sclerotia of A. leporis measure 1000–3000 × 800–1800 μm (Fig. 14D), those of A. aspearensis are 800–1500 × 400–700 μm in size (Fig. 14E) and A. hancockii sclerotia are 500–1200 × 500–800 μm (Fig. 14C).
Fig. 9.
Left to right: 7 d old colonies on CYA, CYA 37 °C, CYA 42 °C, YES, MEA, DG18; top to bottom: A. aflatoxiformans CBS 143679, A. alliaceus CBS 542.65, A. arachidicola CBS 117610, A. aspearensis CBS 143672, A. austwickii CBS 143677, A. avenaceus CBS 109.46, A. bertholletius CBS 143687, A. caelatus CBS 763.97.
Fig. 10.
Left to right: 7 d old colonies on CYA, CYA 37 °C, CYA 42 °C, YES, MEA, DG18; top to bottom: A. cerealis CBS 143674, A. coremiiformis CBS 553.77, A. flavus DTO 258-C9, A. hancockii CBS 142002, A. lanosus CBS 650.74, A. leporis CBS 129235, A. luteovirescens DTO 073-C2 (=NRRL 29235), A. minisclerotigenes DTO 045-F5 (=FRR 4937).
Fig. 11.
Left to right: 7 d old colonies on CYA, CYA 37 °C, CYA 42 °C, YES, MEA, DG18; top to bottom: A. mottae CBS 130016, A. neoalliaceus DTO 326-E7 (=CCF 5413), A. nomius DTO 247-G8, A. novoparasiticus CBS 126849, A. oryzae CBS 100925, A. parasiticus CBS 100926, A. pipericola CBS 143680, A. pseudocaelatus CBS 117616.
Fig. 12.
Left to right: 7 d old colonies on CYA, CYA 37 °C, CYA 42 °C, YES, MEA, DG18; top to bottom: A. pseudonomius CBS 119388, A. pseudotamarii CBS 766.97, A. sergii CBS 130017, A. sojae CBS 100928, A. subflavus CBS 143683, A. tamarii DTO 266-D7, A. togoensis CBS 272.89, A. vandermerwei DTO 368-C2 (= IBT 20468).
Table 5.
Sclerotium and synnema production in species in Aspergillus section Flavi.
Fig. 13.
Sclerotia production by various species belonging to A. flavus-clade. A. A. flavus DTO 281-H8; B. A. flavus DTO 282-A1; C. A. aflatoxiformans CBS 135404; D. A. austwickii CBS 143677; E. A. minisclerotigenes DTO 045-F5; F. A. mottae CBS 130016; G. A. parasiticus DTO 285-G9; H. A. sergii CBS 130017; I. A. subflavus CBS 143683; J. A. cerealis CBS 143675; K. A. pipericola CBS 143680. Scale bar = 500 μm.
Fig. 14.
Sclerotia production by species belonging to Aspergillus section Flavi (and outside the A. flavus-clade; see Fig. 13). A. A. alliaceus CBS 143682; B. A. neoalliaceus CBS 143681; C. A. hancockii CBS 142004; D. A. leporis CBS 129203; E. A. aspearensis CBS 143672; F. A. nomius CBS 260.88; G. A. pseudonomius DTO 267-H7; H. A. caelatus DTO 285-I1; I. A. pseudotamarii CBS 766.97; J. A. bombycis DTO 238-E5. Scale bar = 1000 μm.
Discussion
Mycotoxins and other extrolites
Among the 33 species (including the two domesticated species) in section Flavi, 18 species can produce aflatoxins and one strain of one species, A. bertholletius, can produce the immediate aflatoxin precursor 3-O-methylsterigmatocystin. No fungal species have yet been found that could produce both aflatoxins and ochratoxins. In the A. alliaceus-clade (A. alliaceus, A. neoalliaceus, A. vandermerwei) the conidia are of a yellow shade and these species are able to produce ochratoxin A, but never aflatoxins. Ochratoxin A and B production seem to be an autapomorphy in that clade. On the other hand, two A. alliaceus isolates produced versicolorins (Table 3), which is an intermediate compound in the aflatoxin biosynthetic pathway. This shows that a part of the gene cluster for aflatoxin production may also be present in some species of the A. alliaceus-clade. In the species with yellow-green or brownish green conidia (A. flavus, A. tamarii, A. nomius and A. togoensis clades) several species produce aflatoxins, but never ochratoxins. It is interesting to note that if the ancestor to these five clades produced aflatoxins, then the species in the A. alliaceus clade must have lost the ability to produce aflatoxins, but gained the ability to produce ochratoxins. It has been shown that both ochratoxins and aflatoxins are insecticidal and that kojic acid and aflatoxin are synergistic in insect toxicity (Dowd, 1988, Wicklow et al., 1996). Ochratoxin A and aflatoxin B1 may have similar functions in nature; hence they are never co-produced by any species. It has not been examined whether kojic acid and ochratoxin have a synergistic toxic effect on insects, but kojic acid is produced in large amounts by most species in Aspergillus section Flavi (Varga et al. 2011, Table 3). It should also be noted that aflavinines, aflatrems and aflavazole, found in the sclerotia of many species in section Flavi, are also insecticidal (Gloer et al., 1988, TePaske et al., 1990, TePaske et al., 1992), and thus a number of secondary metabolites from these species may act in concert in repelling insects.
Aspergillus section Flavi contains several species that produce some of the most important mycotoxins known, especially aflatoxins, ochratoxins and cyclopiazonic acid. Eight species are able to produce the B and G type aflatoxins in addition to cyclopiazonic acid: A. aflatoxiformans, A. austwickii, A. cerealis, A. minisclerotigenes, A. mottae, A. pipericola, A. sergii, and A. pseudocaelatus, while A. flavus and A. pseudotamarii produce the B type aflatoxins in addition to cyclopiazonic acid. However, Okoth et al. (2018) found that some of their strains of A. minisclerotigenes produced aflatoxin B only. A. togoensis also produces aflatoxin B1, but not cyclopiazonic acid. A. togoensis is more similar to the aflatoxin B1 producers in Aspergillus subgenus Nidulantes section Ochraceorosei, A. ochraceorosei and A. rambellii, in that all three species accumulate both sterigmatocystin and aflatoxin B1 (Frisvad et al. 2005). These three species have all been isolated from tropical rainforest in the Taï National Forest of Ivory Coast (Bartoli & Maggi 1978), indicating that aflatoxin accumulation pattern is also influenced by the general ecological niches these species occupy. Species producing aflatoxin of the B and G type also include A. nomius, A. luteovirescens and A. novoparasiticus, and of those, A. nomius produces tenuazonic acid in addition to aflatoxins. A. bertholletius is the only species producing both tenuazonic acid and cyclopiazonic acid in addition to an aflatoxin precursor. Species that produce cyclopiazonic acid or tenuazonic acid without producing aflatoxins include A. caelatus, A. tamarii and A. oryzae. The biosynthetic family of cyclopiazonic acids (CPAs) now includes 43 members, including speradines, aspergillines, cyclopiamides and asperorydines (Uka et al., 2017, Liu et al., 2018). Of these 30 members, 22 CPAs have been recovered in A. flavus (Uka et al. 2017). Okoth et al. (2018) reported on cyclopiazonic acid production by an A. parasiticus strain No. 90, and stated that genetic recombination may be the reason for this rare mycotoxin-species connection. Besegmez & Heperkan (2015) also reported on trace CPA production by A. parasiticus strains. We have never observed CPA production in any strain of A. parasiticus or its domesticated form A. sojae. Genome sequencing and annotation of the CPA producing strain No. 90 may help explaining this unexpected result. Since the aflatoxin and CPA gene clusters are neighbours, and CPA is a pathogenicity factor in A. flavus (Chalivandra et al. 2017), A. parasiticus may have the CPA cluster as mostly silent.
It has long been perceived that A. flavus, the most common species in section Flavi, can produce aflatoxin B1 and B2, but not aflatoxin G1 and G2. Here we report on strains of A. flavus sensu stricto from Korea that produce both types of aflatoxin. The only earlier reliable report that A. flavus can produce aflatoxins of the G type was published in 1983 (Wicklow & Shotwell 1983), and it was stated that the G type aflatoxins were only detected in the sclerotia of the genome sequence strain NRRL 3357 (Wicklow and Shotwell, 1983, Nierman et al., 2015). The strains from South Korea are placed in A. flavus both based on phylogeny (Fig. 2, Fig. 3) and extrolite data. Besides the production of aflatoxin G, the extrolite profile of those Korean strains fit well with other A. flavus strains, and including the partially characterized diketopiperazine flavimin that has until now only been found in this species.
Various isolates in section Flavi are able to produce small sclerotia, while those of A. flavus are usually large (Wicklow & Shotwell 1983). Sclerotia in A. flavus and A. parasiticus contain aflatoxins. Furthermore, sclerotium production is associated with specific secondary metabolites, including indoloterpenes such as aflatrems, aflavazole, aflavinines, anominine, aspernomine, paspalines and polyketides such as aflavarins (Table 3; Gallagher and Wilson, 1978, Cole et al., 1981, TePaske et al., 1990, TePaske et al., 1992). Aflatrem was detected in A. aflatoxiformans, A. austwickii, A. cerealis, A. flavus and A. sergii, and aflavazole was detected in A. sergii and A. cerealis (Table 2). The sclerotium associated polyketides aflavarins are also antiinsectan, but has until now only been found in A. flavus (TePaske et al. 1992). The ochratoxin A producing A. alliaceus produce similar isokotanin polyketides in the sclerotia in addition to anominine and paspaline (Gloer et al., 1989, Staub et al., 1993, Laakso et al., 1994), showing the chemical relatedness between A. alliaceus and A. flavus. Aspergillus nomius is also capable of producing anominine in addition to aspernomine (Staub et al., 1992, Bradshaw et al., 2010). The sclerotia of A. leporis and A. aspearensis also contain some aflavinin related metabolites, in addition to unique extrolites (Table 3). While A. hancockii is unique in producing the mycotoxins 7-hydroxytrichothecolone and fumitremorgin A found in other Aspergilli outside section Flavi (Pitt et al. 2017), in general this latter species is chemically unique.
Aspergillus leporis, A. aspearensis and A. hancockii are related species in the Aspergillus leporis clade (Fig. 8). They share few secondary metabolites among them, but do share kojic acid with A. flavus and all other species in Flavi except A. avenaceus and A. coremiiformis. Furthermore, leporine A and other leporines, first found in A. leporis, were later also found in A. flavus (Sun et al., 2014, Arroya-Manzanares et al., 2015, Cary et al., 2015a, Cary et al., 2015b). Aspergillic acid, found in A. flavus (Table 3) and leporins found in A. flavus and A. leporis are strong iron-chelating metabolites. In A. flavus, an aspergillic acid ferri ion complex is readily expressed on the Aspergillus flavus parasiticus agar (AFPA) as an orange reverse, while the leporines are mostly non-expressed (Arroya-Manzanares et al., 2015, Cary et al., 2015a, Cary et al., 2015b) in that species. A. leporis, not being able to produce aspergillic acid, produces leporins more readily. Apart from kojic acid and leporins, A. leporis produces leporizines A, B and C (Reategui et al. 2013). These latter epithiodiketopiperazines are not produced by A. flavus that produces chlorine containing epithiodiketopiperazine heteroisoextrolites instead that are called aspirochlorins (Klausmeyer et al. 2005). Aspirochlorine has been mentioned as a mycotoxin, and has been detected in some strains of A. oryzae, the domesticated form of A. flavus (Monti et al., 1999, Champhamjon et al., 2014).
Occasionally cultures reported to produce new secondary metabolites contain a large number of A. flavus metabolites, and are likely to have been contaminated with A. flavus. For example, Pseudoallescheria boydii F19-1 was reported to produce aflavinine, β-aflatrem, asperfuran, aspergillic acids, cyclopiamide E, 24,25-dehydro-10,11-dihydro-20-hydroxyaflavinine, O-methylsterigmatocystin, pseuboydone E, speradine B and C, in addition to several A. fumigatus metabolites (Lan et al. 2016), so it would be interesting to examine whether the reported pseudoboydones are secondary metabolites from A. flavus or A. fumigatus, and maybe not from P. boydii. In other cases, metabolites from other Aspergilli less closely related species to A. flavus were reported from this species, including terrein, hydroxysydonic acid, gregatin B and aspyrone (Saldan et al. 2018), such data have to be scrutinized and confirmed. We have not been able to detect the latter four secondary metabolites in any strain from Aspergillus section Flavi.
Morphology and ecology
Aspergillus flavus is the most common species in section Flavi causing contamination of food and feed (Klich 2007). The species can be delineated into two major morphotypes: the “L-type”, producing large sclerotia (average diameter >400 μm) and the “S-type”, producing small sclerotia (average diameter <400 μm) (Cotty 1989). In our study we show that A. aflatoxiformans, A. arachidicola, A. austwickii, A. cerealis, A. minisclerotigenes, A. mottae, A. pipericola can produce S-type sclerotia. These species also produce aflatoxins B and G, and strains reported as “strain SBG” can potentially be any of those species (Doster et al., 1996, Freitas-Silva and Vanañcio, 2011, Probst et al., 2007, Probst et al., 2010, Probst et al., 2012, Probst et al., 2014, Wagacha et al., 2013, Arone et al., 2016). The majority of investigated A. flavus strains produce L-type sclerotia, but S-type A. flavus strains occur as well (e.g. NRRL 3251, DTO 281-H8). Contamination events resulting in severe aflatoxicoses in Kenya have been attributed to section Flavi strains that produce S-type sclerotia and B-type aflatoxins (SB). Based on the phylogenetic analysis of nitrate reductase (niaD) and aflatoxin pathway transcription factor (aflR) gene sequences, Probst et al. (2014) hypothese that the Kenyan SB-type isolates comprise a new aflatoxin-producing species. No cultures linked to this outbreak were available in this study for detailed taxonomic analysis. Recent analysis of Eastern Kenyan S-type Flavi strains, isolated from the area experiencing acute aflatoxicosis, showed that these strain are A. flavus or A. minisclerotigenes (Okoth et al. 2018). Based on these data, the unnamed Kenyian SB strain is an A. flavus producing small sized sclerotia. Interestingly, Okoth et al. (2018) also reported A. flavus strains that produced B and G aflatoxins in Eastern Kenya; hence A. flavus SBG also exists. Taken together, A. flavus produces variable sized sclerotia (S or L) and if aflatoxin is produced, then it can be aflatoxin B only or more rarely B and G (SB, SBG or L morphotype).
Some species in section Flavi are widespread and occur foremost in subtropical and tropical climates. A. flavus, A. parasiticus and A. tamarii have been reported from a large number of oil-seeds and nuts (Hedayati et al., 2007, Amaike and Keller, 2011, Varga et al., 2009, Varga et al., 2011, Varga et al., 2015). However, many authors report on the occurrence of other section Flavi species and the presence of other species is therefore more common than first thought. For example, A. minisclerotigenes has been found mostly in South America (Pildain et al. 2008), while A. aflatoxiformans, A. cerealis and A. austwickii, reported as A. flavus SBG, are most common in Africa and Thailand (Probst et al., 2007, Probst et al., 2010, Probst et al., 2012, Probst et al., 2014, Mutegi et al., 2012, Guezlane-Tebibel et al., 2013). Interestingly, many of the (recently) described species producing mycotoxins have been found in foods and are quite common: A. aflatoxiformans (also reported under its synonym A. parvisclerotigenus) (in African corn, Perrone et al., 2014a, Perrone et al., 2014b; Mexican and Nigerian sesame, Ezekiel et al. 2014, this study; edible mushrooms, Ezekiel et al. 2013b; peanut, Frisvad et al. 2005), A. arachidicola (in wild peanuts, Pildain et al. 2008; in Brazil nuts, Gonçalves et al., 2012a, Gonçalves et al., 2012b, Calderari et al., 2013, Taniwaki et al., 2017; in corn, Viaro et al. 2017), A. austwickii (stored rice grains and sesame kernels, this study), A. caelatus (in Brazil nuts, Gonçalves et al., 2012a, Gonçalves et al., 2012b, Taniwaki et al., 2017; in peanuts, Guezlane-Tebibel et al., 2013, Martins et al., 2017), A. cerealis (rice and maize grains, this study; peanut, Carvajal-Campos et al. 2017), A. luteovirescens (in Brazil nuts, Gonçalves et al., 2012a, Gonçalves et al., 2012b, Calderari et al., 2013, Taniwaki et al., 2017), A. nomius (in Brazil nuts, Olsen et al., 2008, Gonçalves et al., 2012a, Gonçalves et al., 2012b, Calderari et al., 2013, Massi et al., 2014, Taniwaki et al., 2017; in cocoa Copetti et al. 2011), A. pseudonomius (in Brazil nuts, Massi et al., 2014, Taniwaki et al., 2017) and to a lesser extent A. novoparasiticus (in corn, Viaro et al. 2017), A. pseudocaelatus (in corn, Viaro et al. 2017; in Brazil nuts, Taniwaki et al. 2017), and A. pseudotamarii (in Brazil nuts, Calderari et al., 2013, Taniwaki et al., 2017). Originally, A. pseudotamarii was found in tea field soil (Ito et al. 2001), A. luteovirescens in silkworm environments (Peterson et al. 2001), A. nomius in bees and in soil and silkworm excrements (Kurtzman et al., 1987, Ito et al., 1998), and A. novoparasiticus as a clinical isolate (Gonçalves et al., 2012a, Gonçalves et al., 2012b). Other species such as A. mottae, A. sergii, A. transmontanensis have been found in corn and almonds in Portugal, but not since their original discovery (Soares et al. 2012). A. togoensis producing sterigmatocystin, aflatoxin B and other secondary metabolites (Wicklow et al., 1989, McAlpin et al., 2000, Rank et al., 2011) has until now only been found on seeds of Landolphia and Strychnos (Samson and Seifert, 1986, Wicklow et al., 1989, Wicklow and McAlpin, 1990). Among the ochratoxin producing species A. alliaceus, A. neoalliaceus and A. vandermerwei, the first species has been detected in onions (Walker & Murphy 1934), peanuts (Wagacha et al. 2013), wheat (Hajjaji et al., 2006, Riba et al., 2008) and tree nuts and figs (Varga et al., 1997, Bayman et al., 2002), while A. neoalliaceus and A. vandermerwei have only been found in soil (Table 1). A. leporis and the species related to it, A. aspearensis and A. hancockii have also only been isolated from soil (States and Christensen, 1966, Christensen, 1981, Varga et al., 2011, Pitt et al., 2017), so even though mycotoxins have been detected such as antibiotic Y in A. leporis (Varga et al. 2011) and a potentially toxic trichothecolone from A. hancockii (Pitt et al. 2017), these species have never been found in foods or feeds. Concerning aflatoxin producers, it is not only A. flavus and A. parasiticus that should be regarded as important producers in foods and feeds, A. aflatoxiformans (= A. parvisclerotigenus), A. arachidicola, A. austwickii, A. luteovirescens, A. cerealis, A. minisclerotigenes, A. nomius, A. novoparasiticus, A. pseudonomius, A. pseudocaelatus and A. pseudotamarii are also aflatoxin producers to be considered.
With exception of A. coremiiformis, all species were able to grow on CYA incubated for 7 d at 37 °C. The majority of species belonging to the A. flavus-clade were able to grow moderate or well at 42 °C (> 5 mm). The only exceptions are A. mottae, A. subflavus (no growth observed) and A. pipericola (CYA42°C (1–5 (–8) mm). Some members of the A. alliaceus-clade were also able to grow at 42 °C, though not consistently (0–8 mm). Growth on creatine agar proved not to be useful to distinguishing species in section Flavi as most species grow poorly on this medium and acid production was not consistent at species level. Some isolates are capable of producing synnemata or synnemata-like structures on Czapek-Dox based media (Bartoli and Maggi, 1978, McAlpin, 2001, Danmek et al., 2014) including A. togoensis, A. caelatus, A coremiiformis and A. flavus, but not A. oryzae, A. nomius, A. parasiticus, and A. pseudotamarii (Danmek et al. 2014). Synnema production has also been reported from tropical rainforest species such as the species Aspergillus dybowskii, A. vitellinus and A. amazonensis (Samson & Seifert 1986), so synnema production may be an ancestral character state in section Flavi. In some of the species, sclerotia are readily formed on most laboratory media, while others are only produced at specific conditions, or only by some isolates. Factors inducing sclerotium formation include corn or corn steep liquor (Wicklow and Shotwell, 1983, Wicklow, 1985, Wicklow and McAlpin, 1990, TePaske et al., 1990, TePaske et al., 1991, TePaske et al., 1992, Rank et al., 2012). In our study we used agar media commonly applied in taxonomic studies investigating Aspergilli (Samson et al. 2014) and we found that sclerotium production most commonly present on CYA incubated at 25 or 37 °C.
Aspergillus section Flavi is the only section in Aspergillus where domesticated species have been accepted as valid species. A. oryzae is the domesticated form of A. flavus, and can be distinguished from the wild type by larger and more smooth conidia having more brown conidium colour en masse, a more floccose colony texture and weaker sporulation, absence of sclerotia, no production of aspergillic acid, and no production of aflatoxins (Wicklow, 1984, Klich and Pitt, 1988, Geiser et al., 2000, Machida et al., 2005, Payne et al., 2006, Hunter et al., 2011, Gibbons et al., 2012). These phenotypical differences may be caused by the interaction of domesticated yeasts (Gibbons & Rinker 2015). Aspergillus sojae is the domesticated form of A. parasiticus, but these two species are morphologically and chemically very similar. Even though A. sojae does not produce aflatoxins, one strain was found to produce versicolorin, which is an aflatoxin precursor.
Taxonomic implications
While most A. flavus strains produce large sclerotia (> 400 μm), some strains uniformly produce small sclerotia (Raper & Fennell, 1965, Hesseltine et al. 1970). Hesseltine et al. (1970) listed NRRL 3251 as an example of a strain with small sclerotia that produced aflatoxin B1 and B2 only. Saito & Tsuruta (1993) studied strains with small sclerotia isolated from agricultural soil in Thailand, including NRRL 3251. They subdivided their strains into two groups: group I produced aflatoxins B1 and B2 (A. flavus SB) and group II produced aflatoxins B1, B2, G1 and G2 (A. flavus SBG). They described their species with small sclerotia as A. flavus var. parvisclerotigenus. This species was typified with NFRI 1538 (SB-type; ex maize field, Chiang Mai, Thailand), but this material is not available for further study. Isolation of Aspergillus section Flavi strains from soil of maize fields in Chiang Mai (Thailand) revealed the presence of strains with small sclerotia (DTO 281-H8) and these strains are identified here as A. flavus. Furthermore, NRRL 3251 belongs, like NFRI 1538T, to group I (Saito & Tsuruta 1993) and also NRRL 3251 is an A. flavus. We therefore treat Aspergillus flavus var. parvisclerotigenus is as a synonym of A. flavus that produces S-type sclerotia and B-type aflatoxins (SB). When Frisvad et al. (2005) raised A. flavus var. parvisclerotigenus to species status as A. parvisclerotigenus, they based it on a neotype from a peanut in Nigeria that produces aflatoxins B1, B2, G1 and G2 (CBS 121.62 = IMI 093070 = NRRL A-11612). The production of small sclerotia in combination with B and G type aflatoxins is linked Saito & Tsuruta's (1993) group II and is therefore in conflict with the protologue of A. flavus var. parvisclerotigenus (Saito & Tsuruta 1993), making the neotypification of A. parvisclerotigenus by Frisvad et al. (2005) incorrect [Art. 9.18 (McNeill et al. 2012)]. This conclusion is also backed up by ecological data because A. parvisclerotigenus sensu Frisvad was not detected in Thai agricultural soils. Actually, A. parvisclerotigenus sensu Frisvad et al. (2005) is mainly found in West Africa: Benin, Burkina Faso, Nigeria, Senegal and Sierra Leone (Probst et al., 2014, Perrone et al., 2014a, Perrone et al., 2014b), but also in Madagascar and from Mexican sesame (this study). Ehrlich et al. (2007) also examined many soil samples in Thailand and found a high number of Aspergillus nomius, a species also producing B and G type aflatoxins, suggesting that group II of Saito & Tsuruta (1993) could be an A. nomius.
Because of the doubtful status of A. parvisclerotigenus sensu Frisvad et al. (2005), we introduce A. aflatoxiformans here for isolates that produce small sclerotia and B and G type aflatoxins, and treat A. parvisclerotigenus as a synonym of A. flavus. Furthermore, three other new species related to A. flavus are introduced (A. austwickii, A. cerealis, A. subflavus), two new species related to A. alliaceus (ochratoxin producers; A. neoalliaceus, A. vandermerwei) and one related to A. leporis (A. aspearensis).
Aspergillus aflatoxiformans Frisvad, Ezekiel, Samson & Houbraken, sp. nov. MycoBank MB823770. Fig. 15.
Fig. 15.
Aspergillus aflatoxiformans CBS 143679T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Sclerotia on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 100 μm; D = 20 μm; E–G = 10 μm.
Etymology: Referring to the copious production of aflatoxins.
Diagnosis: Aspergillus aflatoxiformans is closely related to A. austwickii and A. cerealis, but A. austwickii grows slowly on YES, and Aspergillus cerealis grows slowly on DG18.
Typus: Nigeria, Niger State, Minna, agricultural soil, 2011, collected by C.N. Ezekiel (holotype CBS H-23361, culture ex-type: CBS 143679 = DTO 228-G2 = IBT 32085).
ITS barcode: MG662388. (Alternative markers: BenA = MG517706; CaM = MG518076; RPB2 = MG517897).
Colony diam, 7 d (mm): CYA 50–51; CYA 37 °C 39–40; CYA 42 °C 9–19; MEA 47–50; MEA 37 °C 30–32; MEA 40 °C 22–25; OA 60–70; YES >75; CREA 42–46; CYAS 44–50; DG18 35–38.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse pale luteous (11). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates absent; reverse pale luteous (11). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). CREA 25 °C, 7 d: Growth poor; acid production absent. AFPA: orange reverse.
Micromorphology: Sclerotia 100–250 μm, globose to ellipsoidal, dark brown to black. Conidial heads consistently yellow-green; radiate or loosely columnar, uniseriate. Conidiophores with rough stipes, hyaline, 250–500 × 8–13 μm. Vesicles subglobose to subclavate, 23–38 μm wide, fertile over three fourth of the surface; phialides hyaline, flask-shaped, 7.5–12.5 × 3–5.5 μm. Conidia smooth, subglobose, 3.5–5 × 3–4.5 μm.
Notes: Aspergillus parvisclerotigenus was neotypified with CBS 121.62 (ex Arachis hypogea, Nigeria; Frisvad et al. 2005). This neotypification was incorrect because A. flavus var. parvisclerotigenus originates from Thailand and produces aflatoxin B while A. parvisclerotigenus sensu Frisvad et al. (2005) was neotypified with a strain from Nigeria that produces aflatoxin B and G. Aspergillus flavus var. parvisclerotigenus and A. parvisclerotigenus are placed in synonymy with A. flavus (see also below; List of accepted species and their synonyms in Aspergillus section Flavi) and using the proposed taxonomy, CBS 121.62 is identified as A. aflatoxiformans.
Aspergillus aspearensis Houbraken, Frisvad, Arzanlou & Samson, sp. nov. MycoBank MB823771. Fig. 16.
Fig. 16.
Aspergillus aspearensis CBS 143672T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 20 μm; C–G = 10 μm.
Etymology: Named after Aspear Island (Urmia Lake, Iran), from where the type was isolated.
Diagnosis: Yellow-green, biseriate conidial heads, rough conidiophores, smooth, globose conidia measuring 2.5–3.5 μm.
Typus: Iran, Aspear Island, Urmia Lake, soil, 2012, collected by U. Ghosta & R. Samad (holotype CBS H-23358, culture ex-type: CBS 143672 = DTO 203-D9 = IBT 32590 = IBT 34544).
ITS barcode: MG662398. (Alternative markers: BenA = MG517669; CaM = MG518040; RPB2 = MG517857).
Colony diam, 7 d (mm): CYA 28–70; CYA 37 °C 15–25; CYA 42 °C no growth; MEA 50–65; MEA 37 °C 17–25; OA 50–65; YES >75; CREA 30–40; CYAS 28–65; DG18 45–75.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71); sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense to dense; conidia en masse yellow-green (71); sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71); sclerotia present; soluble pigments absent; exudates absent; reverse buff (45). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse to moderately dense; conidia en masse yellow-green (71); soluble pigments absent; exudates absent; reverse buff (45). OA 25 °C, 7 d: Colonies low, plane; margins entire; mycelium white; texture floccose; sporulation sparse to moderately dense; conidia en masse yellow-green (71); sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). CREA 25 °C, 7 d: Growth poor; acid production absent.
Micromorphology: Sclerotia grey-black, ellipsoidal to irregular, 800–1500 × 400–700 μm. Conidial heads yellow-green; radiate, biseriate. Conidiophores with rough stipes, hyaline, 400–800 × 4.5–7 μm. Vesicles globose, 16–30 μm wide, fertile over the upper half to two thirds; metulae hyaline, 7–9.5 × 3–5 μm; phialides hyaline, flask-shaped, 5.5–8.5 × 2–4 μm. Conidia smooth, globose, 2.5–3.5 μm.
Notes: Aspergillus aspearensis is related to A. leporis and A. hancockii, but produces different secondary metabolites. The only common extrolite between these three species is kojic acid (Table 3).
Aspergillus austwickii Frisvad, Ezekiel, Samson & Houbraken, sp. nov. MycoBank MB823772. Fig. 17.
Fig. 17.
Aspergillus austwickii CBS 143677T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Sclerotia on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 100 μm; D = 20 μm; E–G = 10 μm.
Etymology: Named in honour of Peter K.C. Austwick, a pioneer in the discovery of aflatoxins.
Diagnosis: Aspergillus austwickii is closely related to A. aflatoxiformans and A. cerealis, but A. aflatoxiformans grows faster on YES, and A. cerealis grows slowly on DG18.
Typus: Nigeria, Ogun State, Abeokuta, stored rice grains from market, 2012, collected by C.N. Ezekiel (holotype CBS H-23360, culture ex-type: CBS 143677 = DTO 228-F7 = IBT 32076 = IBT 32590).
ITS barcode: MG662391. (Alternative markers: BenA = MG517702; CaM = MG518072; RPB2 = MG517893).
Colony diam, 7 d (mm): CYA 46–48; CYA 37 °C 37–38; CYA 42 °C 5–20; MEA 45–47; MEA 37 °C 35–37; MEA 40 °C 22–24; OA 60–62; YES 60–65; CREA 40–42; CYAS 46–50; DG18 35–38.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochreous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates absent; reverse pale luteous (11). OA 25 °C, 7 d: Colonies low, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). CREA 25 °C, 7 d: Growth poor; acid production absent. AFPA: orange reverse.
Micromorphology: Sclerotia 100–300 μm, globose to ellipsoidal, dark brown to black. Conidial heads consistently yellow-green; radiate or loosely columnar, uniseriate. Conidiophores with rough stipes, hyaline, 200–500 × 7.5–12.5 μm. Vesicles subglobose to subclavate, 23–33 μm wide, fertile over three fourth of the vesicle surface; phialides hyaline, flask-shaped, 7–10 × 2.5–4.5 μm. Conidia smooth, subglobose, 4–6 × 3.5–5 μm.
Aspergillus cerealis Houbraken, Frisvad, Ezekiel & Samson, sp. nov. MycoBank MB823773. Fig. 18.
Fig. 18.
Aspergillus cerealis CBS 143674T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Sclerotia on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 100 μm; D = 20 μm; E–G = 10 μm.
Etymology: Named based on its occurrence on cereals.
Diagnosis: Aspergillus cerealis is closely related to A. aflatoxiformans and A. austwickii, but A. cerealis grows more slowly on DG18 than the other two species. In addition, A. cerealis is biseriate, while A. aflatoxiformans and A. austwickii are uniseriate. Aspergillus cerealis produces aflavazole, as do some strains of A. flavus, A. minisclerotigenes and A. sergii.
Typus: Nigeria, Ogun State, Shagamu, stored rice grains from market, 2011, collected by C.N. Ezekiel (holotype CBS H-23359, culture ex-type: CBS 143674 = DTO 228-E7 = IBT 32067).
ITS barcode: MG662394. (Alternative markers: BenA = MG517693; CaM = MG518063; RPB2 = MG517884).
Colony diam, 7 d (mm): CYA 60–65; CYA 37 °C 49–51; CYA 42 °C 13–19; MEA 52–55; MEA 37 °C 34–36; MEA 40 °C 18–21; OA 60–63; YES >75; CREA 45–46; CYAS 60–65; DG18 24–27.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44) at centre, pale luteous (11) at edge. DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse pale luteous (11). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). CREA 25 °C, 7 d: Growth poor; acid production absent.
Micromorphology: Sclerotia 100–250 μm, globose to ellipsoidal, dark brown to black. Conidial heads consistently yellow-green; radiate or loosely columnar, biseriate. Conidiophores with rough stipes, hyaline, 1000–2000 × 7–12 μm. Vesicles globose to subglobose, 37–57 μm wide, fertile over entire surface; metulae hyaline, 7–12.5 × 4–6.5 μm; phialides hyaline, flask-shaped, 5–11 × 2.5–4.5 μm. Conidia smooth, subglobose to ellipsoidal, 3–5 × 2.5–4 μm.
Aspergillus neoalliaceus A. Nováková, Hubka, Samson, Frisvad & Houbraken, sp. nov. MycoBank MB823775. Fig. 19.
Fig. 19.
Aspergillus neoalliaceus CBS 143681T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Sclerotia on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 100 μm; D = 20 μm; E–G = 10 μm.
Etymology: Referring to the closely related species Aspergillus alliaceus, but deviating in several features hence A. neoalliaceus.
Diagnosis: Colonies pale to intense yellow when young, turning to cinnamon in age. Conidia smooth, subglobose to ellipsoidal, 2.5–4 × 2–3.5 μm. Sclerotia present.
Typus: Czech Republic, National Reservation Pouzdřanská step, Kolby, soil, 2013, collected by A. Nováková (holotype CBS H-23363, culture ex-type: CBS 143681 = DTO 326-D3 = CCF 5433 = IBT 33110 = IBT 33353).
ITS barcode: MH279420. (Alternative markers: BenA = MG517763; CaM = MG518133; RPB2 = MG517954).
Colony diam, 7 d (mm): CYA 65–75; CYA 37 °C 50–55; CYA 42 °C 0–8; MEA 65–70; MEA 37 °C 43–50; MEA 40 °C 15–18; OA 65–70; YES >75; CREA 55–60; CYAS 65–75; DG18 65–75.
Colony characters: CYA 25 °C, 7 d: Colonies deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse pale luteous (11); soluble pigments absent; exudates present as clear droplets; reverse saffron (10). MEA 25 °C, 7 d: Colonies deep, sulcate; margins entire; mycelium white; texture floccose; sporulation sparse, conidia en masse pale luteous (11) white sclerotia present, turn to dark brown after 10 d; soluble pigments absent; exudates present as clear droplets; reverse sienna (8) at centre, fading into ochreous (44). YES 25 °C, 7 d: Colonies deep, sulcate; margins entire; mycelium white; texture floccose; sporulation absent, large amount of sclerotia present at the edge of colony; soluble pigments absent; exudates present as clear droplets; reverse orange (7) at centre, luteous (12) at edge. DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse luteous (12) at centre, fading into pale luteous (11) or white. OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent, sclerotia present at the edge of colony; soluble pigments absent; exudates present as clear droplets; reverse pale luteous (11). CREA 25 °C, 7 d: Growth poor; acid production absent.
Micromorphology: Sclerotia brownish black, ovate, oblong or oval, 1200–2500 × 800–1200 μm. Conidial heads pale to intense yellow when young, shifting to cinnamon in age; radiate, splitting into columns in age, biseriate. Conidiophores with smooth stipes, hyaline, 2000–3000 × 8.5–13.5 μm. Vesicles globose to subglobose, 40–77 μm wide, fertile over entire surface; metulae hyaline 6.5–11 × 3.5–6 μm; phialides hyaline, flask-shaped, 8–11 × 2–3.5 μm. Conidia smooth, subglobose to ellipsoidal, 2.5–4 × 2–3.5 μm.
Aspergillus pipericola Frisvad, Samson & Houbraken, sp. nov. MycoBank MB823774. Fig. 20.
Fig. 20.
Aspergillus pipericola CBS 143680T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Sclerotia on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 20 μm; D, E = 10 μm; F–G = 10 μm.
Etymology: Referring to pepper, the substrate from which the type was isolated.
Diagnosis: Sporulation is absent on most of media, produces subglobose to elipsoidal conidia measuring 3.5–5.5 × 3.5–5 μm. This species produces small sclerotia and grows restricted at CYA incubated at 42 °C.
Typus: Denmark, black pepper, 2011, collected by J.C. Frisvad (holotype CBS H-23362, culture ex-type: CBS 143680 = DTO 228-H4 = IBT 24628).
ITS barcode: MG662385. (Alternative markers: BenA = MG517717; CaM = MG518087; RPB2 = MG517908).
Colony diam, 7 d (mm): CYA 58–72; CYA 37 °C 70–75; CYA 42 °C 1–5 (–8); MEA 61–72; MEA 37 °C 54–55; OA 52–55; YES >75; CREA 58–65; CYAS 28–30; DG18 62–65.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation absent; dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse cinnamon (62). MEA 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium white; texture floccose; sporulation absent; dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation absent; dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44) to orange (7). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium saffron (10); texture floccose; sporulation sparse; conidia en masse white to pale luteous (11); soluble pigments absent; exudates absent; reverse ochraceous (44) to orange (7). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire white; texture floccose; sporulation absent; dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). CREA 25 °C, 7 d: Growth poor; acid production absent. AFPA: orange reverse.
Micromorphology: Sclerotia 75–250 μm, globose to ellipsoidal, dark brown to black. Conidial heads white to pale luteous; radiate, biseriate. Conidiophores with smooth stipes, hyaline, 900–1200 × 10–16 μm. Vesicles globose, 30–48 μm wide, fertile over entire surface; metulae hyaline, 5.5–8 × 3.5–5 μm; phialides hyaline, flask-shaped, 6–10 × 3.5–5.5 μm. Conidia rough, subglobose to ellipsoidal, 3.5–5.5 × 3.5–5 μm.
Aspergillus subflavus Hubka, A. Nováková, Samson, Frisvad & Houbraken, sp. nov. MycoBank MB823776. Fig. 21.
Fig. 21.
Aspergillus subflavus CBS 143683T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Sclerotia on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 100 μm; D = 20 μm; E–G = 10 μm.
Etymology: The species superficially resembles Aspergillus flavus, hence the name Aspergillus subflavus.
Diagnosis: Colonies yellow-green when young, turn to olive-green in age, uniseriate conidiophores and rough, globose conidia measuring 4.5–6.5 μm. Aspergillus subflavus produces sclerotia that measure 375–650 μm and this species is unable to grow on CYA incubated at 42 °C.
Typus: Romania, above Movile Cave, Dobrogea, Mangalia soil, Sept. 2013, collected by A. Nováková (holotype CBS H-23364, culture ex-type: CBS 143683 = DTO 326-E8 = CCF 4957 = NRRL 66254 = IBT 34939).
ITS barcode: MH279429. (Alternative markers: BenA = MG517773; CaM = MG518143; RPB2 = MG517964).
Colony diam, 7 d (mm): CYA 55–60; CYA 37 °C 15–18; CYA 42 °C No growth; MEA 52–53; MEA 37 °C 7–10; MEA 40 °C No growth; OA 55–60; YES >75; CREA 25–27; CYAS 65–70; DG18 65–75.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse yellow-green (68), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse ochraceous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse yellow-green (71), dark brown sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse pale luteous (11). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse yellow-green (71); soluble pigments absent; exudates absent; reverse buff (45). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse yellow-green (71), sclerotia present; soluble pigments absent; exudates present as clear droplets; reverse buff (45). CREA 25 °C, 7 d: Growth poor; acid production absent.
Micromorphology: Sclerotia 375–650 μm, globose to ellipsoidal, dark brown to black. Conidial heads yellow-green when young, shifting to olive-green in age; loosely radiate, uniseriate. Conidiophores with smooth stipes, hyaline, 300–450 × 7–12.5 μm. Vesicles globose to subglobose, 20–32 μm wide, fertile over three fourth of entire surface; phialides hyaline, flask-shaped, 7.5–13 × 4.5–7 μm. Conidia rough-walled, globose, 4.5–6.5 μm.
Aspergillus vandermerwei Frisvad, Hubka, Samson & Houbraken, sp. nov. MycoBank MB823777. Fig. 22.
Fig. 22.
Aspergillus vandermerwei CBS 612.78T. A. 7 d old colonies: top row left to right, obverse CYA, obverse MEA, YES and OA; bottom row left to right, reverse CYA, reverse MEA, DG18 and CREA. B. Conidial head on MEA. C–F. Conidiophores and conidia. G. Conidia. Scale bars: B = 500 μm; C = 100 μm; D = 50 μm; E–G = 10 μm.
Etymology: Named after K.J. van der Merwe, who contributed to the research on ochratoxin A (Van der Merwe et al. 1965).
Diagnosis: Aspergillus vandermerwei is closely related to A. neoalliaceus, but A. vandermerwei grows slowly on CYA and MEA at 40 °C, and does not produce sclerotia.
Typus: Argentina, Buenos Aires, unknown source, 1950, isolated by J. Winitzky (holotype CBS H-23381, culture ex-type: CBS 612.78 = DTO 069-D2 = DTO 034-B5 = NRRL 5108 = CCF 5683 = IBT 13876).
ITS barcode: EF661567. (Alternative markers: BenA = EF661469; CaM = EF661540; RPB2 = MG517838).
Colony diam, 7 d (mm): CYA 65–73; CYA 37 °C 32–34; CYA 42 °C no growth; MEA 61–68; MEA 37 °C 23–25; MEA 40 °C 1–4; OA 65–75; YES 72–75; CREA 45–50; CYAS 47–50; DG18 53–56.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse pale luteous (11); soluble pigments absent; exudates present as clear droplets; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse pale luteous (11); soluble pigments absent; exudates present as clear droplets; reverse sienna (8) at centre, fading into ochraceous (44). YES 25 °C, 7 d: Colonies moderately dense, sulcate; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates present as clear droplets; reverse pale luteous (11). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse pale luteous (11). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation sparse; conidia en masse pale luteous (11); soluble pigments absent; exudates present as clear droplets; reverse pale luteous (11). CREA 25 °C, 7 d: Growth poor; acid production absent.
Micromorphology: Conidial heads pale to intense yellow when young, shifting to cinnamon in age; radiate, biseriate. Conidiophores with smooth stipes, hyaline, 2000–3000 × 9.5–15.5 μm. Vesicles globose to subglobose, 35–57 μm wide, fertile over entire surface; metulae hyaline 7–8.5 × 3–4.5 μm; phialides hyaline, flask-shaped, 7.5–10 × 2–3.5 μm. Conidia smooth, subglobose to ellipsoidal, 3–4 × 2.5–3.5 μm.
List of accepted species and their synonyms in Aspergillus section Flavi
Below an overview of accepted species in Aspergillus section Flavi (in bold font) and their synonyms. Aspergillus oryzae and A. sojae are domesticated forms of A. flavus and A. parasiticus, respectively. Partial calmodulin gene sequencing, the recommended method for identification of Aspergilli, can’t differentiate these domesticated forms from their wild types. Differentiation between A. oryzae/A. sojae and A. flavus/A. parasiticus is first of all based on the inability of the domesticated forms to produce aflatoxins. The second character for identification is the origin of the strain. Aspergillus oryzae and A. sojae strains are isolated from fermented (food) products or are used in biotechnology. Strains obtained from other environments, even if they are non-aflatoxin producers, are identified as A. flavus. The representatives or ex-type strains of the synonyms listed under A. oryzae and A. sojae were isolated from fermented foods. However, their ability to produce aflatoxins was not studied and this should be done to confirm the proposed classification.
Aspergillus aflatoxiformans Frisvad, Ezekiel, Samson & Houbraken, published here [MB823770]. — Herb.: CBS H-23361. Ex-type: CBS 143679 = DTO 228-G2 = IBT 32085. ITS barcode: MG662388. (Alternative markers: BenA = MG517706; CaM = MG518076; RPB2 = MG517897).
Aspergillus alliaceus Thom & Church, Aspergilli: 163. 1926. [MB256402]. — Herb.: CBS H-7812 (neotype, designated here; MBT 381967). Ex-type: CBS 536.65 = DTO 034-B3 = DTO 046-B1 = ATCC 10060 = DSM 813 = IFO 7538 = IMI 051982 = IMI 051982ii = NRRL 315 = QM 1885 = WB 315. ITS barcode: EF661551. (Alternative markers: BenA = EF661465; CaM = EF661534; RPB2 = MG517825). Notes: Petromyces alliaceus was based on TRTC 46232 (= ATCC 16891 = CBS 542.65 = NRRL 1481; ex soil Australia) and Samson et al. (2014) listed this strain as type of A. alliaceus as well. However, A. alliaceus was based on two strains, one from rotted onions (CBS 110.26 = NRRL 316 = Thom 4660) and the other from a dead blister-beetle (CBS 536.65 = NRRL 315 = Thom 4656; USA) (Thom & Church 1926: 163). NRRL 315 produces a sexual state (Fennell & Warcup 1959) and this strain is therefore selected as neotype of A. alliaceus.
Synonyms: Petromyces alliaceus Malloch & Cain, Can. J. Bot. 50: 2623. 1972. [MB319449]. — Herb.: TRTC 46232. Ex-type: DTO 203-B1 = CBS 542.65 = NRRL 4181 = ATCC 16891 = IMI 126711 = WB 4181. ITS barcode: EF661556. (Alternative markers: BenA = EF661466; CaM = EF661536; RPB2 = EU021644).
Syncleistostroma alliaceum (as ‘alliacea’) Subram., Curr. Sci. 41: 6. 1972. [MB324391]. — Herb.: n/a. Ex-type: CBS 536.65 = DTO 034-B3 = DTO 046-B1 = ATCC 10060 = DSM 813 = IFO 7538 = IMI 051982 = IMI 051982ii = NRRL 315 = QM 1885 = WB 315. ITS barcode: EF661551. (Alternative markers: BenA = EF661465; CaM = EF661534; RPB2 = MG517825).
Aspergillus alliaceus var. macrosterigmatus Glins., Thamavit & Sittir. [nom. inval., Art. 39.1, 40.1 (McNeill et al. 2012)], J. Sci. Soc. Thailand: 43. 1977. [MB347783]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a). Notes: This species was described without a Latin diagnosis and without designation of type material and is therefore invalidly published. This species is tentatively synonymized with A. alliaceus, but could also belong to section Circumdati.
Aspergillus albertensis J.P. Tewari, Mycologia 77: 114. 1985. [MB105069]. — Herb.: UAMH 2976. Ex-type: NRRL 20602 = ATCC 58745 = UAMH 2976. ITS barcode: EF661548. (Alternative markers: BenA = EF661464; CaM = EF661537; RPB2 = EU021628).
Petromyces albertensis J.P. Tewari, Mycologia 77: 114. 1985. [MB105626]. — Herb.: UAMH 2976. Ex-type: NRRL 20602 = ATCC 58745 = UAMH 2976. ITS barcode: EF661548. (Alternative markers: BenA = EF661464; CaM = EF661537; RPB2 = EU021628).
Aspergillus arachidicola Pildain, Frisvad & Samson, Int. J. Syst. Evol. Microbiol. 58: 730. 2008. [MB505189]. — Herb.: unknown. Ex-type: DTO 009-G3 = CBS 117610 = IBT 117610 = IBT 25020. ITS barcode: EF409241. (Alternative markers: BenA = EF203158; CaM = EF202049; RPB2 = MG517802).
Aspergillus aspearensis Houbraken, Frisvad, Arzanlou & Samson, published here [MB823771]. — Herb.: CBS H-23358. Ex-type: CBS 143672 = DTO 203-D9 = CCTU 758 = IBT 32590 = IBT 34544. ITS barcode: MG662398. (Alternative markers: BenA = MG517669; CaM = MG518040; RPB2 = MG517857).
Aspergillus austwickii Frisvad, Ezekiel, Samson & Houbraken, published here [MB823772]. — Herb.: CBS H-23360. Ex-type: CBS 143677 = DTO 228-F7 = IBT 32590 = IBT 32076. ITS barcode: MG662391. (Alternative markers: BenA = MG517702; CaM = MG518072; RPB2 = MG517893).
Aspergillus avenaceus G. Sm., Trans. Brit. Mycol. Soc. 26: 24. 1943. [MB284296]. — Herb.: CBS H-6739. Ex-type: CBS 109.46 = NRRL 517 = ATCC 16861 = IMI 16140 = LCP 89.2592 = LSHBBB 155 = QM 6741 = WB 517. ITS barcode: AF104446. (Alternative markers: BenA = FJ491481; CaM = FJ491496; RPB2 = JN121424).
Aspergillus bertholletius Taniwaki, Pitt & Frisvad, PLoS ONE 7: e42480-P6. 2012. [MB800125]. — Herb.: CCT 7615. Ex-type: DTO 223-D3 = ITAL 270/06 = IBT 29228. ITS barcode: JX198673. (Alternative markers: BenA = MG517689; CaM = JX198674; RPB2 = MG517880).
Aspergillus caelatus B.W. Horn, Mycotaxon 61: 186. 1997. [MB436955]. — Herb.: BPI 737601. Ex-type: DTO 046-A8 = CBS 763.97 = NRRL 25528 = ATCC 201128. ITS barcode: AF004930. (Alternative markers: BenA = EF661470; CaM = EF661522; RPB2 = EF661436).
Aspergillus cerealis Houbraken, Frisvad, Ezekiel & Samson, published here [MB823773]. — Herb.: CBS H-23359. Ex-type: CBS 143674 = DTO 228-E7 = IBT 32067. ITS barcode: MG662394. (Alternative markers: BenA = MG517693; CaM = MG518063; RPB2 = MG517884).
Synonym: Aspergillus korhogoensis A. Carvajal-Campos, A.L. Manizan, S. Tadrist, D.K. Akaki, R. Koffi-Nevry, G.G. Moore, S.O. Fapohunda, S. Bailly, D. Montet, I.P. Oswald, S. Lorber, C. Brabet & O. Puel [nom. inval., art. 42.1 (McNeill et al. 2012)], Toxins 9, 353: 11. 2017. [MB823357]. — Herb.: MACI254. Ex-type: NRRL 66710. ITS barcode: KY689209. (Alternative markers: BenA = KY628792; CaM = KY661267; RPB2 = n/a). Notes: An identifier issued by a recognized repository for that name was not cited in the protologue and this species is therefore not validly described [Art. 42.1 (McNeill et al. 2012)].
Aspergillus coremiiformis Bartoli & Maggi, Trans. Brit. Mycol. Soc. 71: 386. 1979. [MB309214]. — Herb.: RO 102 S. Ex-type: CBS 553.77 = NRRL 13603 = ATCC 38576 = IMI 223069 = NRRL 13756. ITS barcode: EF661544. (Alternative markers: BenA = EU014104; CaM = EU014112; RPB2 = EU021623).
Aspergillus flavus Link, Mag. Ges. Naturf. Freunde Berlin 3: 16, Fr. 1809. [MB209842]. — Herb.: IMI 124930. Ex-type: CBS 569.65 = NRRL 1957 = ATCC 16883 = IMI 124930 = QM 9947 = WB 1957. ITS barcode: AF027863. (Alternative markers: BenA = EF661485; CaM = EF661508; RPB2 = EF661440).
Synonyms: Monilia flava (Link) Pers., Mycol Eur. 1: 30. 1822. [MB496075]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Sterigmatocystis lutea Tiegh (nom. nodum)., Bull. Soc. France 24: 103. 1877. [MB228931]. — Herb.: n/a. Ex-type: CBS 133153 = DTO 214-B2 = WB 508 = NRRL 508 (representative strain, Raper & Fennell 1965: 377). ITS barcode: MH279413. (Alternative markers: BenA = MH279880; CaM = MH279857; RPB2 = n/a). Notes: This species probably served as the basis of Bainier's description of Sterigmatocystis lutea.
Sterigmatocystis lutea Bainier, Bull. Soc. France 27: 30. 1880. [MB219510]. — Herb.: n/a. Ex-type: CBS 133153 = DTO 214-B2 = WB 508 = NRRL 508 (representative strain, Raper & Fennell 1965: 377). ITS barcode: MH279413. (Alternative markers: BenA = MH279880; CaM = MH279857; RPB2 = n/a).
Aspergillus variabilis Gasperini, Atti Soc. Toscana Nat. Sci. Pisa Mem. 8 (Fasc. 1): 326. 1887. [MB161681]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a). Notes: see Aspergillus oryzae var. variabilis.
Sterigmatocystis variabilis (Gasperini) Sacc., Syll. Fung. 10: 525. 1892. [MB197900]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Aspergillus microviridicitrinus Costantin & Lucet, Ann. Sci. Nat. Botan. 2: 158. 1905. [MB535523]. — Herb.: n/a. Ex-type: CBS 124.62 = DTO 067-I8 = IMI 089340 = LSHB BB422 (received at CBS as A. microviridicitrinus). ITS barcode: MH279385. (Alternative markers: BenA = MH279865; CaM = MH279842; RPB2 = n/a).
Aspergillus wehmeri Costantin & Lucet, Ann. Sci. Nat. Botan. (IX) 2: 162. 1905. [MB455472]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Aspergillus effusus Tirab., Ann. Bot. (Rome): 16. 1908. [MB212765]. — Herb.: n/a. Ex-type: CBS 574.65 = DTO 303-C3 = WB 506 = NRRL 506 = ATCC 1010 = IHEM 4388 = IMI 016142 = IMI 124935 = LCP 89.2587 = LSHB Ac21 = NCTC 973 = NRRL 1653 = QM 740 (representative strain, fide Thom and Church, 1926, Thom and Raper, 1945; Raper & Fennell 1965: 377). ITS barcode: JN185448. (Alternative markers: BenA = JN185446; CaM = JN185447; RPB2 = JN185449).
Aspergillus oryzae var. fulvus Yamam. (?), Rept. Govt. Brewing Exptl. Sta. Japan 42. 1912. [MB486957]. — Herb.: n/a. Ex-type: CBS 133118 = DTO 213-I2 = NRRL 4894 = WB 4894 = IMI 359792 (representative, Raper & Fennell 1965: 374). ITS barcode: MH279408. (Alternative markers: BenA = MH279875; CaM = MH279852; RPB2 = n/a). Notes: NRRL 4894 was deposited by the Faculty of Engineering, Osaka University in the NRRL collection as A. oryzae var. fulvus Yamamoto (Wicklow et al. 2002). Raper & Fennell (1965: 374) listed this strain as a representative of A. oryzae var. fulvus and A. flavus var. oryzae f. fulvus.
Aspergillus jeanselmei M. Ota, Annls Parasitol. Humaine Comp.: 146. 1923. [MB268405]. — Herb.: n/a. Ex-type: CBS 108.24 = DTO 389-C1 = NRRL 507 = WB 507 = Thom 5665 (probably ex-type; deposited by M. Ota in the CBS culture collection as A. jeanselmei). ITS barcode: MH279454. (Alternative markers: BenA = MH279882; CaM = MH279859; RPB2 = n/a). No information was found on the source of this species and it is therefore tentatively placed in synonymy with A. flavus.
Sterigmatocystis jeanselmei (N. Ota) Nann., Repertorio sistematico dei miceti dell' uomo e degli animali 4: 229. 1934. [MB252829]. — Herb.: n/a. Ex-type: CBS 108.24 = DTO 389-C1 = NRRL 507 = WB 507 = Thom 5665 (probably ex-type; deposited by M. Ota in the CBS culture collection as A. jeanselmei). ITS barcode: MH279454. (Alternative markers: BenA = MH279882; CaM = MH279859; RPB2 = n/a). Notes: see Aspergillus jeanselmei.
Aspergillus luteus (Tiegh.) C.W. Dodge, Medical mycology. Fungous diseases of men and other mammals: 625. 1935. [MB253119]. — Herb.: n/a. Ex-type: CBS 133153 = DTO 214-B2 = WB 508 = NRRL 508 (representative strain, Raper & Fennell 1965: 365). ITS barcode: MH279413. (Alternative markers: BenA = MH279880; CaM = MH279857; RPB2 = n/a).
Aspergillus flavus var. asper Y. Sasaki, J. Fac. Agric. Hokkaido Imp. Univ. 49: 143. 1950. [MB351898]. — Herb.: n/a. Ex-type: CBS 485.65 = DTO 046-B7 = ATCC 16870 = IFO 5324 = IMI 124932 = LCP 89.3556 = NRRL 4818 = WB 4818 = IBT 3641 = IBT 3657 = JCM 2225 = AHU B-18 (Y. Sasaki). ITS barcode: EF661563. (Alternative markers: BenA = MG517643; CaM = MG518014; RPB2 = MG517828).
Aspergillus thomii G. Sm., Trans. Br. Mycol. Soc. 34: 17. 1951. [MB292861]. — Herb.: n/a. Ex-type: CBS 120.51 = ATCC 16859 = IFO 8135 = IMI 045644 = LCP 56.1517 = LSHB BB213 = NRRL 2097 = NRRL A-2022 = QM 6871 = WB 2097. ITS barcode: EF661549. (Alternative markers: BenA = MG517639; CaM = MG518012; RPB2 = MG517822).
Aspergillus oryzae var. wehmeri (Costantin & Lucet) Y. Ohara, Res. Bull. Fac. Agric., Gifu Univ.: 80. 1953. [MB353278]. — Herb.: n/a. Ex-type: CBS 133063 = DTO 213-H4 = WB 4823 = NRRL 4823 = BCRC 33516 = CCRC 33516 = IAM 2960 = IFO 5770 = JCM 22428 = NBRC 5770 = RIB 1358 = RIFY 5024 = Y. Ohara KK-20 (Ohara's type, Raper & Fennell 1965: 368). ITS barcode: MH279407. (Alternative markers: BenA = MH279874; CaM = MH279851; RPB2 = n/a).
Aspergillus flavus var. microviridicitrinus (Costantin & Lucet) Nehira, J. Ferment. Technol., Osaka 35: 56. 1957. [MB500159]. — Herb.: n/a. Ex-type: CBS 124.62 = DTO 067-I8 = IMI 089340 = LSHB BB422 (received at CBS as A. microviridicitrinus). ITS barcode: MH279385. (Alternative markers: BenA = MH279865; CaM = MH279842; RPB2 = n/a).
Aspergillus flavus var. oryzae f. fulvus (Yamam.{?}) Nehira, J. Ferment. Technol., Osaka 35: 56. 1957. [MB347785]. — Herb.: n/a. Ex-type: CBS 133118 = DTO 213-I2 = NRRL 4894 = WB 4894 = IMI 359792 (representative, Raper & Fennell 1965: 374). ITS barcode: MH279408. (Alternative markers: BenA = MH279875; CaM = MH279852; RPB2 = n/a). Notes: see Aspergillus oryzae var. fulvus.
Aspergillus flavus var. proliferans Anguli, Rajam, Thirum., Rangiah & Ramamurthi [nom. inval., Art. 39.1 (McNeill et al. 2012)], Indian Journal of Microbiology 5: 94. 1965. [MB349038]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Aspergillus subolivaceus Raper & Fennell, Gen. Aspergillus: 385. 1965. [MB326661]. — Herb.: IMI 44882. Ex-type: CBS 501.65 = DTO 046-B5 = NRRL 4998 = ATCC 16862 = IMI 44882 = NRRL 20625 = QM 8902 = WB 4998. ITS barcode: AF257795. (Alternative markers: BenA = MG517642; CaM = MG518015; RPB2 = MG517827).
Aspergillus flavus var. columnaris Raper & Fennell, Gen. Aspergillus: 366. 1965. [MB349037]. — Herb.: WB 4818. Ex-type: CBS 485.65 = DTO 046-B7 = ATCC 16870 = IFO 5324 = JCM 2225 = IMI 124932 = LCP 89.3556 = NRRL 4818 = WB 4818 = IBT 3641 = IBT 3657 = AHU B-18 (Y. Sasaki). ITS barcode: EF661563. (Alternative markers: BenA = MG517643; CaM = MG518014; RPB2 = MG517828).
Aspergillus flavus var. parvisclerotigenus Mich. Saito & Tsuruta, Proc. Jpn. Assoc. Mycotoxicol. 37: 32. 1993. [MB361049]. — Herb.: NFRI 1538. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a). Notes: The original type culture and herbarium specimen of A. flavus var. parvisclerotigenus is unavailable. Strains with the same phenotype (small sclerotia and aflatoxin B production) are identified as A. flavus and this species is therefore considered as a synonym of A. flavus.
Aspergillus parvisclerotigenus (Mich. Saito & Tsuruta) Frisvad & Samson, Syst. Appl. Microbiol., 28: 450. 2005. [MB500166]. — Herb.: CBS 121.62 (neotype). Ex-type: DTO 223-C2 = CBS 121.62 = IMI 93070 = NRRL A-11612 = IBT 3651. ITS barcode: EF409240. (Alternative markers: BenA = MG517683; CaM = MG518054; RPB2 = MG517874). Notes: The original type culture and herbarium specimen of A. flavus var. parvisclerotigenus is unavailable (Frisvad et al. 2005) and it was therefore neotypified with CBS 121.62 (ex Arachis hypogea, Nigeria). Aspergillus flavus var. parvisclerotigenus originates from Thailand and produces aflatoxin B while A. parvisclerotigenus sensu Frisvad et al. (2005) was neotypified with a strain from Nigeria that produces aflatoxin B and G. The neotypification of A. parvisclerotigenus is therefore incorrect (this study). Aspergillus parvisclerotigenus sensu Frisvad et al. is in this study described as a new species named Aspergillus aflatoxiformans.
Petromyces flavus B.W. Horn, I. Carbone & G.G. Moore, Mycologia 101: 424. 2009. [MB512910]. — Herb.: BPI 878851. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a). Notes: The holotype of Petromyces flavus is a dried slant culture of A. flavus NRRL 29473 (MAT1-1) crossed with A. flavus NRRL 29478 (MAT1-2) that produces cleistothecia and ascospores.
Aspergillus hancockii Pitt PLoS ONE e0170254: 16. 2017. [MB818219]. — Herb.: FRR 3425. Ex-type: CBS 142004 = DTO 360-G7. ITS barcode: KX858342. (Alternative markers: BenA = MBFL01001228.1:26000-28000; CaM = MBFL01000377.1:5000-7000; RPB2 = MBFL01000137:9000-11000).
Aspergillus lanosus Kamal & Bhargava, Trans. Brit. Mycol. Soc. 52: 336. 1969. [MB326640]. — Herb.: IMI 130727. Ex-type: CBS 650.74 = DTO 034-B7 = NRRL 3648 = IMI 130727 = QM 9183 = WB 5347. ITS barcode: EF661553. (Alternative markers: BenA = MG517633; CaM = MG518017; RPB2 = EU021642).
Aspergillus luteovirescens Blochwitz, Ann. Mycol. 31 (1-2): 80. 1933. [MB269992]. — Herb.: CBS H-23401 (neotype, designated here; MBT 381966). Ex-type: CBS 620.95 = DTO 010-H1 = CBS 116.32 (dead) = IMI 348034 = NRRL 4858 = WB 4858. ITS barcode: MG662406. (Alternative markers: BenA = MG517625; CaM = MG517998; RPB2 = MG517808).
Synonym: Aspergillus bombycis S.W. Peterson, Yoko Ito, B.W. Horn & T. Goto, Mycologia 93: 691. 2001. [MB474687]. — Herb.: BPI 745225. Ex-type: CBS 117187 = DTO 046-B8 = NRRL 26010 = IBT 23536 = IMI 386978 = NBRC 100700. ITS barcode: AF104444. (Alternative markers: BenA = AY017547; CaM = AY017594; RPB2 = EF661458).
Aspergillus leporis States & M. Chr., Mycologia 58: 738. 1966. [MB326641]. — Herb.: NY RMF 99. Ex-type: CBS 151.66 = NRRL 3216 = ATCC 16490 = NRRL A-14256 = NRRL A-15810 = QM 8995 = RMF99 = WB 5188. ITS barcode: AF104443. (Alternative markers: BenA = EF661499; CaM = EF661541; RPB2 = EF661459).
Aspergillus minisclerotigenes Vaamonde, Frisvad & Samson, Int. J. Syst. Evol. Microbiol. 58: 733. 2008. [MB505188]. — Herb.: unknown. Ex-type: CBS 117635 = DTO 009-F7 = DTO 303-C6 = IBT 25032. ITS barcode: EF409239. (Alternative markers: BenA = EF203148; CaM = MG518009; RPB2 = MG517799).
Aspergillus mottae C. Soares, S.W. Peterson & Venâncio, Mycologia 104: 692. 2012. [MB561841]. — Herb.: MUM-H 10.231. Ex-type: CBS 130016 = DTO 223-C8. ITS barcode: JF412767. (Alternative markers: BenA = HM803086; CaM = MG518058; RPB2 = MG517878).
Aspergillus neoalliaceus A. Nováková, Hubka, Samson, Frisvad & Houbraken, published here [MB823775]. — Herb.: CBS H-23363. Ex-type: CBS 143681 = DTO 326-D3 = S765 = CCF 5433 = IBT 33110 = IBT 33353. ITS barcode: MH279420. (Alternative markers: BenA = MG517763; CaM = MG518133; RPB2 = MG517954).
Aspergillus nomius Kurtzman et al., Antonie van Leeuwenhoek 53: 151. 1987. [MB133392]. — Herb.: BPI NRRL 13137. Ex-type: CBS 260.88 = NRRL 13137 = ATCC 15546 = FRR 3339 = IMI 331920 = LCP 89.3558 = NRRL 6108 = NRRL A-13671 = NRRL A-13794. ITS barcode: AF027860. (Alternative markers: BenA = AF255067; CaM = AY017588; RPB2 = EF661456).
Synonyms: Aspergillus zhaoqingensis Z.T. Qi & Z.M. Sun, Acta Mycol. Sin.: 22. 1991. [MB130300]. — Herb.: HMAS 58980. Ex-type: CBS 399.93 = DTO 301-I8 = AS 3.4626. ITS barcode: FJ491472. (Alternative markers: BenA = MG517757; CaM = MG518127; RPB2 = MG517948).
Petromyces nomius B.W. Horn, I. Carbone & G.G. Moore, Mycologia 103: 176. 2011. [MB518289]. — Herb.: BPI 880386. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a). Notes: The holotype of Petromyces nomius is a dried slant culture of A. nomius NRRL 26886 (MAT1-1/MAT1-2) crossed with A. nomius NRRL 58994 (MAT1-2) that produces cleistothecia and ascospores.
Aspergillus novoparasiticus S.S. Gonçalves, Stchigel, Cano, Godoy-Martinez, Colombo & Guarro, Med. Mycol. 50: 158. 2011. [MB516612]. — Herb.: CBS H-20401. Ex-type: CBS 126849 = DTO 223-C3 = LEMI 250 = FMR 10121. ITS barcode: MG662397. (Alternative markers: BenA = MG517684; CaM = MG518055; RPB2 = MG517875).
Aspergillus oryzae (Ahlb.) Cohn, Jahresber. Schles. Ges. Vaterl. Cult. 61: 226. 1884. [MB184394]. — Herb.: IMI 16266. Ex-type: CBS 100925 = CBS 102.07 = NRRL 447 = ATCC 1011 = ATCC 12891 = ATCC 4814 = ATCC 7561 = ATCC 9102 = IAM13118 = IFO 4075 = IFO 537 = IFO 5375 = IMI 16266 = IMI 44242 = LSHBA c .19 = NCTC 598 = NRRL 692 = QM 6735 = Thom 113 = WB 447. ITS barcode: EF661560. (Alternative markers: BenA = EF661483; CaM = EF661506; RPB2 = EF661438).
Synonyms: Eurotium oryzae Ahlb., Dingler's Polytechn. J.: 330. 1878. [MB225012]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Aspergillus pseudoflavus Saito, Centbl. Bakt. ParasitKde, Abt. 2 18: 34. 1907. [MB188103]. — Herb.: n/a. Ex-type: CBS 133059 = DTO 213-F2 = WB 4787 = NRRL 4787 = IMI 360437 = IFO 4083 = JCM 2066 = IAM 2956 = ATU, A-68-6 (representative; Raper & Fennell 1965: 375, Wicklow et al. 2002). ITS barcode: MH279402. (Alternative markers: BenA = MH279869; CaM = MH279846; RPB2 = n/a). Notes: NRRL 4787 is one of K. Saito’s strains upon which Ohara (1953) based his recognition of A. oryzae var. pseudoflavus (Saito) Ohara (Raper & Fennell 1965: 375). This strain was isolated from fermented food and therefore identified here as A. oryzae. The blue-green pigmentation reported by Saito for old colonies has been observed in strain NRRL 483 (= CBS 132943 = DTO 213-C8 = WB 483 = IMI 360438 = Thom 3526) (Raper & Fennell 1965: 376). NRRL 483 was isolated by Wehmer (before 1914; data NRRL culture collection) and is according Raper & Fennell (1965: 376) a representative of A. pseudoflavus. However, the source of Wehmer's strain is probably not a fermented food product and this strain is therefore tentatively identified as A. flavus.
Aspergillus gymnosardae Yukawa, J. Coll. Agric. Imp. Univ. Tokyo: 362. 1911. [MB167015]. — Herb.: n/a. Ex-type: CBS 114.32 = DTO 067-H4 = QM 9703 (received as A. gymnosardae at CBS, originating from Japan). ITS barcode: MH279399. (Alternative markers: BenA = MH279866; CaM = MH279843; RPB2 = n/a). Notes: WB 505 (= CBS 132941 = DTO 213-C6 = NRRL 505) is another representative of this species and was received from Japan. Aspergillus gymnosardae was reported as essential to the ripening of the tuna fish preparation, “katsuobushi” (Raper & Fennell 1965:373).
Sterigmatocystis pseudoflava (Saito) Sacc., Syll. Fung. 22: 1260. 1913. [MB194870]. — Herb.: n/a. Ex-type: CBS 133059 = DTO 213-F2 = WB 4787 = NRRL 4787 = IMI 360437 = IFO 4083 = JCM 2066 = IAM 2956 = ATU, A-68-6 (representative; Raper & Fennell 1965: 375, Wicklow et al. 2002). ITS barcode: MH279402. (Alternative markers: BenA = MH279869; CaM = MH279846; RPB2 = n/a). Notes: see Aspergillus pseudoflavus.
Aspergillus oryzae var. globosus Sakag. & K. Yamada, J. Agric. Chem. Soc. Japan 20: 72. 1944. [MB351901]. — Herb.: n/a. Ex-type: CBS 133107 = DTO 214-A9 = WB 5004 = NRRL 5004 = IMI 359789 = IFO 4242 = IAM 2667 = NBRC 4242 = RIB 1301 = JCM 2242 = K. Sakaguchi SH 10-5. ITS barcode: MH279411. (Alternative markers: BenA = MH279878; CaM = MH279855; RPB2 = n/a). Notes: NRRL 5004 was isolated from rom Shoyu-koji, Chiba Prefecture, Japan and represents A. oryzae var. globosus and A. flavus var. oryzae f. globosus.
Aspergillus oryzae var. magnasporus Sakag. & K. Yamada, J. Agric. Chem. Soc. Japan 20: 72. 1944. [MB346544]. — Herb.: n/a. Ex-type: CBS 133158 = DTO 214-B7 = WB 4804 = NRRL 4804 = JCM 22379 = IAM 2673 = Sakaguchi strain SH-8-4 (representative, Raper & Fennell 1965: 366). ITS barcode: MH279414. (Alternative markers: BenA = MH279881; CaM = MH279858; RPB2 = n/a). Notes: CBS 133158 was isolated from Shoyu-koji in Japan and this strain is a representative of A. oryzae var. magnasporus (Raper & Fennell 1965: 366).
Aspergillus oryzae var. microsporus Sakag. & K. Yamada, J. Agric. Chem. Soc. Japan 20: 73. 1944. [MB346545]. — Herb.: n/a. Ex-type: CBS 133108 = DTO 214-B1 = NRRL 5003 = WB 5003 = IMI 359796 = IFO 4233 = K. Sakaguchi A5-1 (representative, Raper & Fennell 1965:374). ITS barcode: MH279412. (Alternative markers: BenA = MH279879; CaM = MH279856; RPB2 = n/a).
Aspergillus candidus var. amylolyticus Takaoka [nom. inval. Art. 39.1 (McNeill et al. 2012)], J. Agr. Chem. Soc. Japan 23:57. 1949. [MB493812]. — Herb.: n/a. Ex-type: CBS 466.91 = DTO 389-C8 = NRRL 5032 = IFO 6215 = WB 5032 = IMI 360440. ITS barcode: MH279451. (Alternative markers: BenA = MH279886; CaM = MH279862; RPB2 = n/a). Notes: This species produces white coloured conidia. No information was found on the source of this species but this species is generally accepted as A. oryzae (Raper & Fennell 1965).
Aspergillus oryzae var. effusus (Tirab.) Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 81. 1951. [MB123955]. — Herb.: n/a. Ex-type: CBS 133112 = DTO 213-I7 = WB 5030 = NRRL 5030 = IMI 360436 = IFO 5321 (representative of Ohara’s A. oryzae var. effusus; Raper & Fennell 1965: 377, Wicklow et al. 2002). ITS barcode: MH279409. (Alternative markers: BenA = MH279876; CaM = MH279853; RPB2 = n/a). Notes: NRRL 5030 is the basis for I. Ohara’s recognition of A. oryzae var. effusus (Wicklow et al. 2002). According Raper & Fennell (1965: 377), Ohara’s strain (NRRL 5030 = CBS 133112) differs from the original description of A. effusus (represented by NRRL 506 = CBS 574.65). CBS 574.65 was isolated from Zea mays from Vermont, USA and based on ecology and sequence data this strain is identified as A. flavus. CBS 133112 was isolated from fermented food and is therefore identified as A. oryzae.
Aspergillus oryzae var. pseudoflavus (Saito) Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 81. 1951. [MB349041]. — Herb.: n/a. Ex-type: CBS 133059 = DTO 213-F2 = WB 4787 = NRRL 4787 = IMI 360437 = IFO 4083 = JCM 2066 = IAM 2956 = ATU, A-68-6 (representative; Raper & Fennell 1965: 375, Wicklow et al. 2002). ITS barcode: MH279402. (Alternative markers: BenA = MH279869; CaM = MH279846; RPB2 = n/a). Notes: see Aspergillus pseudoflavus.
Aspergillus oryzae var. sporoflavus Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 81. 1951. [MB349042]. — Herb.: n/a. Ex-type: CBS 133064 = DTO 213-E8 = WB 4824 = NRRL 4824 = IAM 2957 = IFO 5785 = JCM 2067 = NBRC 5785 = RIB 1366 = Y. Ohara, MM-1-1 (Raper & Fennell 1965: 368). ITS barcode: MH279401. (Alternative markers: BenA = MH279868; CaM = MH279845; RPB2 = n/a). Notes: NRRL 4824 was isolated from miso-koji in Japan and represents A. oryzae var. sporoflavus (Raper & Fennell 1965: 368).
Aspergillus oryzae var. microvesiculosus Y. Ohara, J. Agric. Chem. Soc. Japan 26: 550. 1952. [MB346546]. — Herb.: n/a. Ex-type: CBS 133042 = DTO 213-F4 = WB 4803 = NRRL 4803 = IMI 359794 = IFO 4203 = IAM 2633 = JCM 2233 = JCM 2246 = NBRC 4203 = RIB 1160 = K. Sakaguchi M 1–2 (representative, Raper & Fennell 1965: 375). ITS barcode: MH279403. (Alternative markers: BenA = MH279870; CaM = MH279847; RPB2 = n/a). Notes: NRRL 4803 was listed as a representative of A. oryzae var. microvesiculosus (Raper & Fennell 1965: 375). This strain was isolated from koji for miso, Kumamoto Prefecture by Prof. Ken-ichiro Sakaguchi, The University of Tokyo and is therefore identified as A. oryzae.
Aspergillus oryzae var. tenuis Y. Ohara, J. Agric. Chem. Soc. Japan 26: 550. 1952. [MB351902]. — Herb.: n/a. Ex-type: CBS 133044 = DTO 213-G1 = WB 4799 = NRRL 4799 = IMI 359791 = IFO 4134 = CCRC 31251 = IAM 2601 = IAM 2958 = IHEM 5780 = JCM 10114 = JCM 2068 = JCM 22426 = NBRC 4134 = RIB 1362 = RIB 3010. ITS barcode: MH279404. (Alternative markers: BenA = MH279871; CaM = MH279848; RPB2 = n/a). Notes: NRRL 4799 belongs to T. Takahashi’s “A. oryzae – D,” and was isolated from koji for sake, and is the basis for A. oryzae var. tenuis (Wicklow et al. 2002).
Aspergillus sojae var. gymnosardae (Yukawa) Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 77. 1953. [MB349044]. — Herb.: n/a. Ex-type: CBS 133045 = DTO 213-G4 = WB 4806 = NRRL 4806 = IMI 360439 = IFO 4294 = NBRC 4294 = JCM 2226 (representative; Raper & Fennell 1965:376, Wicklow et al. 2002). ITS barcode: MH279405. (Alternative markers: BenA = MH279872; CaM = MH279849; RPB2 = n/a). Notes: WB 4806 (= CBS 133045 = DTO 213-G4 = NRRL 4806 = IMI 360439 = IFO 4294 = NBRC 4294 = JCM 222) was isolated from katsuobushi (dried bonito) and is the basis for I. Ohara’s recognition of A. sojae var. gymnosardae (Raper & Fennell 1965:376, Wicklow et al. 2002). See also notes of A. gymnosardae.
Aspergillus flavus var. oryzae f. magnasporus (Sakag. & K. Yamada) Nehira, J. Ferment. Technol., Osaka 35: 56. 1957. [MB347787]. — Herb.: n/a. Ex-type: CBS 133158 = DTO 214-B7 = WB 4804 = NRRL 4804 = JCM 22379 = IAM 2673 = Sakaguchi strain SH-8-4 (representative, Raper & Fennell 1965: 366). ITS barcode: MH279414. (Alternative markers: BenA = MH279881; CaM = MH279858; RPB2 = n/a). Notes: See Aspergillus oryzae var. magnasporus.
Aspergillus oryzae var. variabilis (Gasperini) Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 84. 1953. [MB346548]. — Herb.: n/a. Ex-type: CBS 133062 = DTO 213-G6 = IAM 2959 = IFO 5768 = JCM 2247 = NBRC 5768 = NRRL 4822 = QM 8892 = RIB 1364 = WB 4822 = Y. Ohara, KK-9 (representative strain). ITS barcode: EF661564. (Alternative markers: BenA = EF661490; CaM = EF661513; RPB2 = EF661445). Notes: NRRL 4822 fails to conform to Gasperini's (and Ohara's) description (fide Raper & Fennell 1965: 368) and it's questionable whether this strain is a good representative of A. oryzae var. variabilis and A. variabilis.
Aspergillus flavus var. oryzae f. globosus (Sakag. & K. Yamada) Nehira, J. Ferment. Technol., Osaka 35: 56. 1957. [MB347786]. — Herb.: n/a. Ex-type: CBS 133107 = DTO 214-A9 = WB 5004 = NRRL 5004 = IMI 359789 = IFO 4242 = IAM 2667 = NBRC 4242 = RIB 1301 = JCM 2242 = K. Sakaguchi SH 10-5. ITS barcode: MH279411. (Alternative markers: BenA = MH279878; CaM = MH279855; RPB2 = n/a). Notes: See Aspergillus oryzae var. globosus.
Aspergillus flavus var. oryzae f. microsporus (Sakag. & K. Yamada) Nehira, J. Ferment. Technol., Osaka 35: 56. 1957. [MB347788]. — Herb.: n/a. Ex-type: CBS 133108 = DTO 214-B1 = NRRL 5003 = WB 5003 = IMI 359796 = IFO 4233 = K. Sakaguchi A5-1 (representative, Raper & Fennell 1965: 374). ITS barcode: MH279412. (Alternative markers: BenA = MH279879; CaM = MH279856; RPB2 = n/a).
Aspergillus parasiticus f. gymnosardae (Yukawa) Nehira, J. Ferment. Technol., Osaka 35: 56. 1957. [MB347794]. — Herb.: n/a. Ex-type: CBS 114.32 = DTO 067-H4 = QM 9703 (received as A. gymnosardae at CBS, originating from Japan). ITS barcode: MH279399. (Alternative markers: BenA = MH279866; CaM = MH279843; RPB2 = n/a). Notes: see Aspergillus gymnosardae.
Aspergillus oryzae var. brunneus Murak., J. Gen. Appl. Microbiol. (Tokyo) 17: 304. 1971. [MB352617]. — Herb.: RIB 1172. Ex-type: CBS 817.72 = DTO 389-C2 = IHEM 4381 = MUCL 31309 = IAM 2648 = IFO 30102 = JCM 2240 = K. Sakaguchi, S-3-8, ACTU 0-10-8. ITS barcode: MH279453. (Alternative markers: BenA = MH279883; CaM = MH279860; RPB2 = n/a). Notes: Isolated from sake-koji, Japan.
Aspergillus oryzae var. viridis (as ”viride”) Murak., J. Gen. Appl. Microbiol. (Tokyo) 17: 303. 1971. [MB352619]. — Herb.: RIB 128. Ex-type: CBS 819.72 = DTO 389-D2 = ATCC 22788 =IFO 30113 =IHEM 4382 = JCM 2248 = MUCL 31310 = VTT D-88355. ITS barcode: MH279450. (Alternative markers: BenA = MH279887; CaM = MH279863; RPB2 = n/a). Notes: this species was described from sake-koji, Japan.
Aspergillus flavus subsp. flavus var. oryzae (Ahlb.) Kurtzman, M.J. Smiley, Robnett & Wicklow, Mycologia 78: 957 1986. [MB130238]. — Herb.: n/a. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Aspergillus parasiticus Speare, Bull. Div. Pathol. Physiol., Hawaiian Sugar Planters Assoc. Exp. Sta. 12: 38. 1912. [MB191085]. — Herb.: IMI 15957ix. Ex-type: CBS 100926 = CBS 103.13 = NRRL 502 = ATCC 1018 = ATCC 6474 = ATCC 7865 = IMI 15957 = IMI 15957ii = IMI 15957iv = IMI 15957ix = IMI 15957vi = IMI 15957vii = LCP 89.2566 = LSHBA c 14 = NCTC 975 = NRRL 1731 = NRRL 3315 = NRRL A-13360 = NRRL A-14693 = Thom 3509 = WB 502. ITS barcode: AY373859. (Alternative markers: BenA = EF661481; CaM = AY017584; RPB2 = EF661449).
Synonyms: Aspergillus terricola var. americanus Marchal, Am. J. Bot. 8: 125. 1921. [MB124083]. — Herb.: WB 424. Ex-type: CBS 580.65 = DTO 046-B9 = ATCC 1014 = ATCC 16863 = IMI 016127 = IMI 016127ii = LSHB Ac22 = NCTC 974 = NRRL 424 = QM 7475 = VKM F-2041 = WB 424. ITS barcode: MG662404. (Alternative markers: BenA = MG517644; CaM = MG518030; RPB2 = MG517829).
Aspergillus chungii Y.K. Shih, Lingnan Sci. J.: 365. 1936. [MB251412]. — Herb.: n/a. Ex-type: CBS 115.37 = DTO 303-C2 = NRRL 4868 = IMI 093122 = WB 4868. ITS barcode: FJ491464. (Alternative markers: BenA = MG517759; CaM = MG518129; RPB2 = MG517950).
Aspergillus parasiticus var. globosus Murak., J. Gen. Appl. Microbiol. (Tokyo) 12: 195. 1966. [MB353279]. — Herb.: ATCC 15517. Ex-type: CBS 260.67 = DTO 046-C2 = ATCC 15517 = CCM F-550 = CECT 2680 = DSM 2038 = IFO 30179 = IHEM 4387 = IMI 120920 = IMI 229041 = MUCL 31311. ITS barcode: MG662400. (Alternative markers: BenA = EF203156; CaM = MG518013; RPB2 = MG517830).
Aspergillus toxicarius Murak., J. Gen. Appl. Microbiol. (Tokyo) 17: 307. 1971. [MB309247]. — Herb.: IMI 089717. Ex-type: CBS 822.72 = DTO 046-A9 = DTO 389-C9 = ATCC 22789 = IFO 30109 = IMI 089717 = RIB 4002 =TPI M 39. ITS barcode: MG662401. (Alternative markers: BenA = EF203163; CaM = MG518019; RPB2 = MG517824).
Aspergillus flavus subsp. parasiticus var. parasiticus (Speare) Kurtzman, M.J. Smiley, Robnett & Wicklow, Mycologia 78: 958. 1986. [MB130237]. — Herb.: IMI 15957ix. Ex-type: CBS 100926 = CBS 103.13 = NRRL 502 = ATCC 1018 = ATCC 6474 = ATCC 7865 = IMI 15957 = IMI 15957ii = IMI 15957iv = IMI 15957ix = IMI 15957vi = IMI 15957vii = LCP 89.2566 = LSHBA c 14 = NCTC 975 = NRRL 1731 = NRRL 3315 = NRRL A-13360 = NRRL A-14693 = Thom 3509 = WB 502. ITS barcode: AY373859. (Alternative markers: BenA = EF661481; CaM = AY017584; RPB2 = EF661449).
Aspergillus americanus (Marchal & É.J. Marchal) Kozak., Mycol. Pap. 161: 163. 1989. [MB127757]. — Herb.: . Ex-type: CBS 580.65 = DTO 046-B9 = ATCC 1014 = ATCC 16863 = IMI 016127 = IMI 016127ii = LSHB Ac22 = NCTC 974 = NRRL 424 = QM 7475 = VKM F-2041 = WB 424. ITS barcode: MG662404. (Alternative markers: BenA = MG517644; CaM = MG518030; RPB2 = MG517829).
Petromyces parasiticus B.W. Horn, I. Carbone & J.H. Ramirez-Prado, Mycologia 101: 276. 2009. [MB513282]. — Herb.: BPI 878821. Ex-type: n/a. ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a). Notes: The holotype of Petromyces parasiticus is a dried slant culture of A. parasiticus NRRL 29538 (MAT1-1) crossed with A. parasiticus NRRL 29570 (MAT1-2) that produces cleistothecia and ascospores.
Aspergillus pipericola Frisvad, Samson & Houbraken, published here [MB823774]. — Herb.: CBS H-23362. Ex-type: CBS 143680 = DTO 228-H4 = IBT 24628. ITS barcode: MG662385. (Alternative markers: BenA = MG517717; CaM = MG518087; RPB2 = MG517908).
Aspergillus pseudocaelatus Varga, Samson & Frisvad, Stud. Mycol. 69: 63. 2011. [MB560397]. — Herb.: CBS H-20632. Ex-type: CBS 117616 = DTO 010-H4. ITS barcode: EF409242. (Alternative markers: BenA = MG517626; CaM = MG517995; RPB2 = MG517809).
Aspergillus pseudonomius Varga, Samson & Frisvad, Stud. Mycol. 69: 67. 2011. [MB560398]. — Herb.: CBS H-20633. Ex-type: CBS 119388 = DTO 009-F1 = NRRL 3353 = IBT 27864. ITS barcode: AF338643. (Alternative markers: BenA = EF661495; CaM = EF661529; RPB2 = EF661454).
Aspergillus pseudotamarii Yoko Ito, S.W. Peterson, Wicklow & T. Goto, Mycol. Res. 105: 237. 2001. [MB466527]. — Herb.: BPI 746098. Ex-type: CBS 766.97 = DTO 046-C1 = NRRL 25517. ITS barcode: AF272574. (Alternative markers: BenA = EF203125; CaM = EF202030; RPB2 = EU021631).
Aspergillus sergii P. Rodrigues, S.W. Peterson, Venâncio & N. Lima, Mycologia 104: 693. 2012. [MB561842]. — Herb.: MUM-H 10.219. Ex-type: CBS 130017 = DTO 223-C9 = DTO 223-D1. ITS barcode: JF412769. (Alternative markers: BenA = MG517688; CaM = MG518059; RPB2 = HM802985).
Aspergillus sojae Sakag. & K. Yamada, J. Agric. Chem. Soc. Japan 20: 72. 1944. [MB102834]. — Herb.: IMI 191300. Ex-type: CBS 100928 = DTO 046-C3 = IMI 191300. ITS barcode: KJ175434. (Alternative markers: BenA = KJ175494; CaM = KJ175550; RPB2 = MG517831).
Synonym: Aspergillus flavus subsp. parasiticus var. sojae (Sakag. & K. Yamada ex Murak.) Kurtzman, M.J. Smiley, Robnett & Wicklow, Mycologia 78: 958. 1986. [MB130239]. — Herb.: IMI 191300. Ex-type: CBS 100928 = DTO 046-C3 = IMI 191300. ITS barcode: KJ175434. (Alternative markers: BenA = KJ175494; CaM = KJ175550; RPB2 = MG517831).
Aspergillus subflavus Hubka, A. Nováková, Samson, Frisvad & Houbraken, published here [MB823776]. — Herb.: CBS H-23364. Ex-type: CBS 143683 = DTO 326-E8 = S778 = CCF 4957 = NRRL 66254 = IBT 34939. ITS barcode: MH279429. (Alternative markers: BenA = MG517773; CaM = MG518143; RPB2 = MG517964).
Aspergillus tamarii Kita, Centralbl. Bakteriol. 2. Abth. 37: 433. 1913. [MB191425]. — Herb.: CBS 104.13. Ex-type: CBS 104.13 = NRRL 20818 = QM 9374. ITS barcode: AF004929. (Alternative markers: BenA = EF661474; CaM = EF661526; RPB2 = EU021629).
Synonyms: Aspergillus terricola É.J. Marchal, Revue Mycol. (Toulouse): 101. 1893. [MB191770]. — Herb.: IMI 172294. Ex-type: CBS 579.65 = ATCC 16860 = IMI 172294 = NRRL 426 = WB 426. ITS barcode: EF661559. (Alternative markers: BenA = EF661472; CaM = EF661525; RPB2 = EU021649). Notes: The name Aspergillus terricola competes with Aspergillus tamarii. The former species has priority based on publication date (1893 vs 1913). Marchal's (1893) description of A. terricola is incomplete and he describes the A. terricola as strictly uniseriate (Raper & Fennell 1965). Although this character can vary on different media and culture ages, it remains questionable whether Marchal was dealing with an A. tamarii. Because no type is known to be preserved of A. terricola, the species was neotypified with IMI 172294 (= CBS 579.65 = ATCC 16860 = NRRL 426 = WB 426) (Samson & Gams 1985). In contrast, the lectotype culture of A. tamarii CBS 104.13 (ex koji, Japan) was received at CBS from G. Kita. Based on these data, the identity of A. terricola is unclear while A. tamarii is unambiguously defined with an original culture.
Aspergillus flavus mut. rufa Blochwitz, Ann. Mycol. 27: 196. 1929 [MB123823]. — Herb.: n/a. Ex-type: n/a (Raper & Fennell 1965: 384). ITS barcode: n/a. (Alternative markers: BenA = n/a; CaM = n/a; RPB2 = n/a).
Aspergillus lutescens Bainier ex Thom & Raper, A manual of the Aspergilli: 251. 1945. [MB284305]. — Herb.: NRRL 425. Ex-type: NRRL 425 = QM 7418 = Thom 4640.478. ITS barcode: EF661558. (Alternative markers: BenA = EF661475; CaM = EF661524; RPB2 = EU021648).
Aspergillus terricola var. bronzeus Saincl., Centralbl. Gesammte Forstwesen: 118. 1949. [MB351905]. — Herb.: n/a. Ex-type: CBS 129.49 = DTO 389-C6. ITS barcode: KJ175440. (Alternative markers: BenA = MH279884; CaM = KJ175555; RPB2 = n/a).
Aspergillus parasiticus var. rugosus Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 78. 1953. [MB353280]. — Herb.: n/a. Ex-type: CBS 133375 = DTO 389-C7 = WB 4960 = NRRL 4960. ITS barcode: MH279452. (Alternative markers: BenA = MH279885; CaM = MH279861; RPB2 = n/a).
Aspergillus tamarii var. crassus Y. Ohara, Res. Bull. Fac. Agric. Gifu Univ.: 76. 1953. [MB353282]. — Herb.: n/a. Ex-type: CBS 133097 = DTO 213-H5 = NRRL 4959 = WB 4959. ITS barcode: MG662403. (Alternative markers: BenA = MG517678; CaM = MG518049; RPB2 = MG517866).
Aspergillus effusus var. furcatus Bat. & H. Maia, Anais Soc. Biol. Pernambuco 13: 93. 1955. [MB351896]. — Herb.: DMUR 8. Ex-type: CBS 133104 = DTO 214-A6 = WB 4910 = NRRL 4910 = IMI 360444. ITS barcode: MH279410. (Alternative markers: BenA = MH279877; CaM = MH279854; RPB2 = n/a).
Aspergillus flavofurcatus Bat. & H. Maia, Anais Soc. Biol. Pernambuco 13: 94. 1955. [MB292844]. — Herb.: DMUR 318. Ex-type: CBS 484.65 = NRRL 4911 = ATCC 16864 = IHEM 4385 = IMI 124938 = LCP 89.2591 = MUCL 31304 = WB 4911. ITS barcode: EF661565. (Alternative markers: BenA = EF661473; CaM = EF661527; RPB2 = EU021651).
Aspergillus indicus B.S. Mehrotra & Agnihotri, Mycologia 54: 403. 1963. [MB326637]. — Herb.: Allahabad A-29. Ex-type: CBS 167.63 = DTO 010-G9 = NRRL 4680 = ATCC 15054 = IMI 172295 = QM 8903 = WB 4680. ITS barcode: MG662407. (Alternative markers: BenA = MG517624; CaM = MG518001; RPB2 = MG517807).
Aspergillus terricola var. indicus (B.S. Mehrotra & Agnihotri) Raper & Fennell, Gen. Aspergillus: 412. 1965. [MB353283]. — Herb.: Allahabad A-29. Ex-type: CBS 167.63 = DTO 010-G9 = NRRL 4680 = ATCC 15054 = IMI 172295 = QM 8903 = WB 4680. ITS barcode: MG662407. (Alternative markers: BenA = MG517624; CaM = MG518001; RPB2 = MG517807).
Aspergillus togoensis (Henn.) Samson & Seifert, Adv. Penicillium Aspergillus Syst.: 419. 1985. [MB114720]. — Herb.: BR B 1009. Ex-type: CBS 205.75 = NRRL 13551 = LCP 67.3456 (CBS 272.89 = DTO 034-C1 (representative strain). ITS barcode: AJ874113. (Alternative markers: BenA = FJ491477; CaM = FJ491489; RPB2 = JN121479).
Aspergillus transmontanensis P. Rodrigues, S.W. Peterson, N. Lima & Venâncio, Mycologia 104: 694. 2012. [MB561843]. — Herb.: MUM-H 10.214. Ex-type: DTO 223-C7 = CBS 130015. ITS barcode: JF412774. (Alternative markers: BenA = HM803101; CaM = HM803020; RPB2 = HM802980).
Aspergillus vandermerwei Frisvad, Hubka, Samson & Houbraken, published here [MB823777]. — Herb.: CBS H-23381. Ex-type: CBS 612.78 = DTO 069-D2 = DTO 034-B5 = NRRL 5108 = CCF 5683 = IBT 13876. ITS barcode: EF661567. (Alternative markers: BenA = EF661469; CaM = EF661540; RPB2 = MG517838).
Chemical synoptic key for Aspergillus section Flavi
Species list
1 A. aflatoxiformans
2 A. alliaceus
3 A. arachidicola
4 A. aspearensis
5 A. austwickii
6. A. avenaceus
7. A. bertholletius
8. A. caelatus
9 A. cerealis
10 A. coremiiformis
11 A. flavus
12 A. hancockii
13 A. lanosus
14 A. leporis
15 A. luteovirescens
16 A. minisclerotigenes
17 A. mottae
18 A. neoalliaceus
19 A. nomius
20 A. novoparasiticus
21 A. oryzae
22 A. parasiticus
23 A. pipericola
24 A. pseudocaelatus
25 A. pseudonomius
26 A. pseudotamarii
27 A. sergii
28 A. sojae
29 A. subflavus
30 A. tamarii
31 A. togoensis
32 A. transmontanensis
33 A. vandermerwei
Aflatoxin B type: 1, 3, 5, 9, 11, 15, 16, 17, 19, 20, 22, 23, 24, 25, 26, 27, 31, 32
Aflatoxin G type: 1, 3, 5, 9, (11), 15, 16, 17, 19, 20, 22, 23, 24, 25, 27, 32
Aflatrem: 1, 5, 9, 11, 16, 23, 27
Aflavarins, isokotanins, kotanins, siderins: 1, 2, 5, 9, 11, 12, 16, 23, 27, 33
Aflavazol: 9, 11, 16, 27
Aflavinines: 1, 4, 11, 16, 17, (21), 23, 26, 27, 29
“Alkca”: 8, 24, 26
Altersolanols: 2, 6, 8, 13, 15, 26, 33
Anominine: 2, 18, 19, 33
Antarone A: 2
Antibiotic Y: 14
Asperfuran: 11, 21, 27, 28
Aspernomin: 19
Asperopterin*: 21
Aspergillic acid: 1, 3, 5, 9, 11, 15, 16, 17, 19, 20, 22, 23, 24, 26, 27, 28, 32
Aspergillomarasmines*: 11, 21
Aspirochlorin: 1, 6, 8, 11, 20, 21, 24, 26, 28, 29, 30, 32, 33
Asperlicin: 2, 13, 33
Brefeldin A: 18, 33
Chrysogine: 3, 15, 26, 28
Citreoisocoumarin: 11, 21, (30)
Clavatols: 14
Cyclopiazonic acid: 1, 5, 7, 9, 11, (12, speradine F), 16, 17, 21, 23, 24, 26, 27, 30
Dehydroterrestric acid: 12
Ditryptoleucine: 21
Ditryptophenaline: 3, 11, 20, 24
Eupenifeldin*: 12
Flavimin (not structure elucidated): 11
Fumaryl-D,L-alanine*: 30
“Gfn”: 1, 5
Griseofulvin: 13, (33)
Hancockiamides: 12
7-Hydroxytrichothecolone: 12
Kojic acid: 1, 2, 3, 4, 5, 7, 8, 9, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 32, 33
Leporins: 11, 14
Leporizines: 14
“Met I”: 2, 13
Mevinolin: 4
Miyakamides / oryzamides: 3, 11, 15, 19, 20, 21, 26, 28, 32
3-Nitropropionic acid*: 6, 11, 21, 30
Ochratoxins: 2, 18, 33
Onycins: 12
Parasiticolides/astelollides: 3, 7, 11, 16, 21, 22, 29
Paspaline, paspalinine: 1, 2, 4, 5, 9, 11, 14, 16, 17, 18, 19, (21), 23, 26, 27, 31
Paxillin: 31
Penicillins*: 11, 21, 22
Pseurotin A: (6), 14, 19
Sporogen AO1: 15, 21
Tenuazonic acid: 7, 8, 15, 19, 24, 25, 26, 30
“Tetracyclic compound”: 20
Ustilaginoidin C: 7, 11, 22
Versicolorins: 1, (2), 3, 5, (7), 9, 11, 15, 16, 17, 19, 20, 22, 23, 24, 25, 26, 27, (28), 31, 32
*Note: The strains in this study were not screened for these extrolites. The data are based on literature and only isolates with verified identity are included.
Acknowledgements
This project was supported by CAMS Innovation Fund for medical Sciences (CIFMS) 2017-12M-1-013 and by the National Key Research and Development Program of China (No. 2017ZX09101003-006-006) and by National Science Foundation of China No. 81473345. The Czech contributions were supported by the project BIOCEV (CZ.1.05/1.1.00/02.0109) provided by the Ministry of Education, Youth and Sports of CR and ERDF and Charles University Research Centre program No. 204069. JCF and TOL thank the Novo Nordic Foundation for support via the grant NNF13OC0005201. JCF thanks Agilent for an Agilent Thought Leader Award and support from the Danish National Research Foundation (DNRF137) for the Center for Microbial Secondary Metabolites. S-BH thanks the support of the Rural Development Administration in Korea (project number PJ01354902) and JH thanks Martin Meijer and Bart Kraak for their assistance. The authors thank Iamtaweejaroen Panrapee and Anukul Nampeung for their help on the isolation and characterisation of strains from Thai soils and Sofia Chulze for thanked for sharing cultures used in this study. The advice of Konstanze Bensch on nomenclature issues is greatly acknowledged.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.simyco.2018.06.001.
Contributor Information
J.C. Frisvad, Email: jcf@bio.dtu.dk.
J. Houbraken, Email: j.houbraken@westerdijkinstitute.nl.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Adler M., Wintersteiner O. A reinvestigation of flavacidin, the penicillin produced by Aspergillus flavus. Journal of Biological Chemistry. 1948;176:873–891. [PubMed] [Google Scholar]
- Amaike S., Keller N.P. Aspergillus flavus. Annual Review of Phytopathology. 2011;49:107–133. doi: 10.1146/annurev-phyto-072910-095221. [DOI] [PubMed] [Google Scholar]
- Amare M.G., Keller N.P. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genetics and Biology. 2014;66:11–18. doi: 10.1016/j.fgb.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Ammar H.A.M., Srour A.Y., Ezzat S.M. Identification and characterization of genes involved in kojic acid biosynthesis in Aspergillus flavus. Annals of Microbiology. 2017;67:691–702. [Google Scholar]
- Arnstein H.R.V., Cook A.H. The penicillin produced by Aspergillus parasiticus. British Journal of Experimental Pathology. 1947;28:94–98. [PMC free article] [PubMed] [Google Scholar]
- Arone L., Augusto J., Bandyopahyay R. Diversity of Aspergillus section Flavi S morphotype in Mozambique. Phytopathology. 2016;106:24. [Google Scholar]
- Arroya-Manzanares N., Di Mavungu D., Uka V. Use of UHPLC high resolution Orbitrap mass spectrometry to investigate the genes involved in the production of secondary metabolites in Aspergillus flavus. Food Additives and Contaminants. Part A – Chemistry Analysis Control Exposure & Risk Assessment. 2015;32:1656–1673. doi: 10.1080/19440049.2015.1071499. [DOI] [PubMed] [Google Scholar]
- Arzanlou M., Samadi R., Frisvad J.C. Two novel Aspergillus species from hypersaline soils of The National Park of Lake Urmia, Iran. Mycological Progress. 2016;15:1081–1092. [Google Scholar]
- Asai Y., Nonaka N., Nishio M. TMC-2A, -2B, and -2C, new dipeptidyl peptidase inhibitors produced by Aspergillus oryzae A374. II. Isolation and structure determination. Journal of Antibiotics. 1998;50:653–658. doi: 10.7164/antibiotics.50.653. [DOI] [PubMed] [Google Scholar]
- Assante G., Camarda L., Locci R. Isolation and structure of red pigments from Aspergillus flavus and related species, grown on a differential medium. Journal of Agricultural and Food Chemistry. 1981;29:785–787. [Google Scholar]
- Atlas R.M. CRC Press; Boca Raton: 2010. Handbook of Microbiological Media. [Google Scholar]
- Baker J.L., Bayman P., Mahoney N.E. Proceedings of the 3rd Fungal Genomics, 4th fumonisin, and 16th aflatoxin elimination workshop, Savannah, Georgia. 2003. Ochratoxigenic Aspergillus lanosus and A. alliaceus from California tree nut orchards. [Google Scholar]
- Barayani N., Despot D.J., Palagyi A. Identification of Aspergillus species in central Europe able to produce G-type aflatoxins. Acta Biologica Hungarica. 2015;66:339–347. doi: 10.1556/018.66.2015.3.9. [DOI] [PubMed] [Google Scholar]
- Barbier M., Vetter W., Bogdanov D. Synthese und Eigenschaften eines Analogen des Lycomarasmins und der Aspergillomarasmine. Annalen der Chemie-Justus Liebig. 1963;668:132. [Google Scholar]
- Bartoli A., Maggi O. 4 new species of Aspergillus from Ivory Coast. Transactions of the British Mycological Society. 1978;71:383–394. [Google Scholar]
- Basaran P., Demirbas R.M. Spectroscopic detection of pharmaceutical compounds from an aflatoxigenic strain of Aspergillus parasiticus. Microbiological Research. 2010;165:516–522. doi: 10.1016/j.micres.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Bayman P., Baker J.L., Doster M.A. Ochratoxin A production by the Aspergillus ochraceus group and Aspergillus alliaceus. Applied and Environmental Microbiology. 2002;68:2326–2329. doi: 10.1128/AEM.68.5.2326-2329.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G.E., Schmidt E.L. β-nitropropionic acid and nitrite in relation to nitrate formation by Aspergillus flavus. Archives of Microbiology. 1964;49:167–175. [PubMed] [Google Scholar]
- Berg D.H., Massing R.P., Hoehn M.M. A30641, a new epidithiodiketopiperazine with antifungal activity. Journal of Antibiotics. 1976;29:394–397. doi: 10.7164/antibiotics.29.394. [DOI] [PubMed] [Google Scholar]
- Besegmez H.I.O., Heperkan D. Aflatoxin, cyclopiazonic acid and beta-nitropropionic acid production by Aspergillus section Flavi from dried figs grown in Turkey. Quality Assurance and Safety of Crops and Foods. 2015;7:477–485. [Google Scholar]
- Birch A.J., Qureshi A.A., Rickards R.W. Metabolites of Aspergillus indicus: The structure and some aspects of the biosynthesis of dihydrocanadensolide. Australian Journal of Chemistry. 1968;21:2775–2784. [Google Scholar]
- Birkinshaw J.H., Charles J.H.V., Lilly C.H. The biochemistry of microorganisms. VII. Kojic acid (5-hydroxy-m-methylpyrone) Philosophical Transactions of the Royal Society, London. 1931;B220:127–138. [Google Scholar]
- Blinc M., Johanides V. Antibiotics from aspergilli with special regard to species isolated in Yugoslavia. Bulletin Science, Conseil Academie RPF Yugoslavie. 1956;2:99. [Google Scholar]
- Bradshaw B., Etxebarria-Jardí G., Bonjoch J. Total synthesis of (-)anominine. Journal of the American Chemical Society. 2010;132:5966–5967. doi: 10.1021/ja101994q. [DOI] [PubMed] [Google Scholar]
- Brookes D., Tidd B.K., Turner W.B. Avenaciolide, an antifungal lactone from Aspergillus avenaceus. Journal of the Chemical Society. 1963;1963:5385–5391. [Google Scholar]
- Brown D.W., Hauser F.M., Tommasi R. Structural elucidation of a conidial pigment from Aspergillus parasiticus. Tetrahedron Letters. 2003;34:419–422. [Google Scholar]
- Buchanan R.L., Ayres J.C. Effect of sodium acetate on growth and aflatoxin production by Aspergillus parasiticus NRRL 2999. Journal of Food Science. 1976;41:128–132. [Google Scholar]
- Büchi G., Francisco M.A., Murray W.V. Aspersitin – a new metabolite of Aspergillus parasiticus. Tetrahedron Letters. 1983;24:2527–2530. [Google Scholar]
- Bush M., Goth A. Flavicin: an antibacterial substance produced by Aspergillus flavus. Journal of Pharmacology and Experimental Therapy. 1943;78:164–169. [Google Scholar]
- Bush M., Goth A., Dickison H.L. Flavicin II: An antibacterial substance produced by an Aspergillus flavus. Journal of Pharmacology and Experimental Therapy. 1945;84:262–277. [Google Scholar]
- Bush M., Touster O., Brockman E. The production of β-nitropropionic acid by a strain of Aspergillus flavus. Journal of Biological Chemistry. 1951;188:685–693. [PubMed] [Google Scholar]
- Calderari T.O., Iamanaka B.T., Frisvad J.C. The biodiversity of Aspergillus section Flavi in brazil nuts: from rainforest to producer. International Journal of Food Microbiology. 2013;160:267–272. doi: 10.1016/j.ijfoodmicro.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Camiletti B.X., Torrico A.K., Fernando Maurino M. Fungal screening and aflatoxin production by Aspergillus section Flavi isolated from pre-harvest maize ears grown in two Argentinean regions. Crop Protection. 2017;92:41–48. [Google Scholar]
- Cardwell K.F., Cotty P.J. Distribution of Aspergillus section Flavi among soils from the four agricultural zones of the republic of Bénin, West Africa. Plant Disease. 2002;86:434–439. doi: 10.1094/PDIS.2002.86.4.434. [DOI] [PubMed] [Google Scholar]
- Carvajal-Campos A., Manizan A.L., Tadriest S. Aspergillus korhogoensis, a novel aflatoxin producing species from Côte d’Ivoire. Toxins. 2017;9:353. doi: 10.3390/toxins9110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J.W., Harris-Coward P.Y., Ehrlich K.C. Functional characterization of a veA-dependant polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genetics and Biology. 2014;64:25–35. doi: 10.1016/j.fgb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Cary J.W., Uka V., Han Z. An Aspergillus flavus secondary metabolite gene cluster containing a hybrid PKS-NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genetics and Biolology. 2015;81:88–97. doi: 10.1016/j.fgb.2015.05.010. [DOI] [PubMed] [Google Scholar]
- Cary J.W., Han Z., Yin Y. Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryotic Cell. 2015;14:983–997. doi: 10.1128/EC.00092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J.W., Harris-Coward P., Scharfenstein L. The Aspergillus flavus homeobox gene, hbx1, is required for development and aflatoxin production. Toxins. 2017;9:315. doi: 10.3390/toxins9100315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalivandra S.C., DeRobertis C., Chang P.-K. Cyclopiazonic acid is a pathogenicity factor for Aspergillus flavus and a promising target for screening germplasm for ear rot resistance. Molecular Plant-Microbe Interactions. 2017;30:361–373. doi: 10.1094/MPMI-02-17-0026-R. [DOI] [PubMed] [Google Scholar]
- Champhamjon P., Boettger-Schmidt D., Scherlach K. Biosynthesis of the halogenated mycotoxin aspirochlorine in koji mold involves cryptic amino acid conversion. Angewandte Chemie International Edition. 2014;53:13409–13413. doi: 10.1002/anie.201407624. [DOI] [PubMed] [Google Scholar]
- Chang P.-K., Ehrlich K.C. Cyclopiazonic acid biosynthesis by Aspergillus flavus. Toxin Reviews. 2011;30:79–89. [Google Scholar]
- Chang P.-K., Horn B.W., Dorner J.W. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthetic gene cluster in Aspergillus flavus. Fungal Genetics and Biology. 2009;46:176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Chang P.-K., Scharfenstein L.L., Li R.W. Aspergillus flavus aswA, a gene homolog of Asperguillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genetics and Biology. 2017;104:29–37. doi: 10.1016/j.fgb.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Christensen C.M., Nelson G.H., Speers G.M. Results of feeding tests with rations containing grain invaded by a mixture of naturally present fungi plus Aspergillus flavus NRRL 2999. Minnesota research suggests that danger of toxicity from material invaded by a mixture of fungi probably is not very great. Feedstuffs. 1973;45:20–41. [Google Scholar]
- Christensen M. A synoptic key and evaluation of species in the Aspergillus flavus group. Mycologia. 1981;73:1056–1084. [Google Scholar]
- Ciegler A. Bioproduction of ochratoxin A and penicillic acid by members of the Aspergillus ochraceus group. Canadian Journal of Microbiology. 1972;18:631–636. doi: 10.1139/m72-100. [DOI] [PubMed] [Google Scholar]
- Codner R.C., Sargeant K., Yeo R. Production of aflatoxin by the culture of strains of Aspergillus flavus-oryzae on sterilized peanuts. Biotechnology and Bioengineering. 1963;5:185–192. [Google Scholar]
- Cole R.J., Dorner J.W., Springer J.P. Indole metabolites from a strain of Aspergillus flavus. Journal of Agricultural and Food Chemistry. 1981;29:293–295. [Google Scholar]
- Copetti M.V., Iamanaka B.T., Pereira J.L. Aflatoxigenic fungi and aflatoxins in cocoa. International Journal of Food Microbiology. 2011;148:141–144. doi: 10.1016/j.ijfoodmicro.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Cotty P.J. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology. 1989;79:808–814. [Google Scholar]
- Cotty P.J. Influence of field application of an atoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton balls and on the aflatoxin content of cottonseed. Phytopathology. 1994;84:1270–1277. [Google Scholar]
- Cotty P.J., Cardwell K.F. Divergence of West African and north American communities of Aspergillus section Flavi. Applied and Environmental Microbiology. 1999;65:2264–2266. doi: 10.1128/aem.65.5.2264-2266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danmek K., Prasongsuk S., Lotrakul P. Effect of Avid (R) on the synnema-like formation of Aspergillus flavus grown on Czapek medium. African Journal of Microbiology Research. 2014;5:2812–2815. [Google Scholar]
- Donner M., Atenkeng J., Sikora R.A. Distribution of Aspergillus section Flavi in soils of maize fields in three agri–ecological zones of Nigeria. Soil Biology and Biochemistry. 2009;41:37–44. [Google Scholar]
- Dorner J.W. Production of cyclopiazonic acid by Aspergillus tamarii Kita. Applied and Environmental Microbiology. 1983;46:1435–1437. doi: 10.1128/aem.46.6.1435-1437.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doster M., Michailides T., Morgan D. Aspergillus species and mycotoxins in figs from California orchards. Plant Disease. 1996;80:484–489. [Google Scholar]
- Dowd P.F. Synergism of aflatoxin B1 toxicity with co-occurring fungal metabolite kojic acid to 2 caterpillars. Entomologia Experimentalis et Applicata. 1988;47:69–71. [Google Scholar]
- Doxtater K.G., Alexander M. Role of 3-nitropropionic acid in nitrate formation by Aspergillus flavus. Journal of Bacteriology. 1966;91:186–191. doi: 10.1128/jb.91.3.1186-1191.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K.C. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K.C., Mack B.M. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins. 2014;6:1916–1928. doi: 10.3390/toxins6061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K.C., Kobbeman K., Montalbo B.G. Aflatoxin-producing Aspergillus species from Thailand. International Journal of Food Microbiology. 2007;114:153–159. doi: 10.1016/j.ijfoodmicro.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Ezekiel C.N., Sulyok M., Babalola D.A. Incidence and consumer awareness of toxigenic Aspergillus section Flavi and aflatoxin B1 in peanut cake from Nigeria. Food Control. 2013;30:596–601. [Google Scholar]
- Ezekiel C.N., Sulyok M., Frisvad J.C. Fungal and mycotoxin assessment of dried edible mushroom in Nigeria. International Journal of Food Microbiology. 2013;162:231–236. doi: 10.1016/j.ijfoodmicro.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Ezekiel C.N., Udom I.E., Frisvad J.C. Assessment of aflatoxigenic Aspergillus and other fungi in millet and sesame from Plateau State, Nigeria. Mycology. 2014;5:16–22. doi: 10.1080/21501203.2014.889769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustinelli P.C., Palencia E.R., Sobole V.S. Study of the genetic diversity of the aflatoxin biosynthetic cluster in Aspergillus section Flavi using insertion/deletion markers in peanut seeds from Georgia, USA. Mycologia. 2017;109:200–209. doi: 10.1080/00275514.2017.1307095. [DOI] [PubMed] [Google Scholar]
- Faustinelli P.C., Wang X.M., Palencia E.R. Genome sequences of eight Aspergillus flavus spp. and one A. parasiticus sp., isolated from peanut seeds in Georgia. Genome Announcements. 2016;4:e00278–e00316. doi: 10.1128/genomeA.00278-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N.D., Khaldi N., Joarder V.S. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell D.I., Warcup J.H. The ascocarps of Aspergillus alliaceus. Mycologia. 1959;51:409–415. [Google Scholar]
- Filtenborg O., Frisvad J.C., Svendsen J.A. Simple screening method for moulds producing intracellular mycotoxins in pure cultures. Applied and Environmental Microbiology. 1983;45:581–585. doi: 10.1128/aem.45.2.581-585.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Silva O., Vanañcio A. Brazil nuts: benefits and risks associated with contamination by fungi and mycotoxins. Food Research International. 2011;44:1434–1440. [Google Scholar]
- Frisvad J.C. Media and growth conditions for induction of secondary metabolites. In: Keller N.P., Turner G., editors. Fungal secondary metabolism: methods and protocols. Vol. 944. Humana Press; New York: 2012. pp. 47–58. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Larsen T.O., de Vries R. Secondary metabolite profiling, growth profiles and other tools for species recognition and important Aspergillus mycotoxins. Studies in Mycology. 2007;59:31–37. doi: 10.3114/sim.2007.59.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Samson R.A. Neopetromyces gen. nov. and an overview of teleomorphs of Aspergillus subgenus Circumdati. Studies in Mycology. 2000;45:201–207. [Google Scholar]
- Frisvad J.C., Skouboe P., Samson R.A. Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Systematic and Applied Microbiology. 2005;28:442–453. doi: 10.1016/j.syapm.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Thrane U. Standardized High-Performance Liquid Chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone indices and UV-VIS spectra (diode-array detection) Journal of Chromatography. 1987;404:195–214. doi: 10.1016/s0021-9673(01)86850-3. [DOI] [PubMed] [Google Scholar]
- Galagan J.E., Calvo S.E., Cuomo C. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gallagher R.T., Wilson B.J. Aflatrem, the tremorgenic mycotoxin from Aspergillus flavus. Mycopathologia. 1978;66:183–185. doi: 10.1007/BF00683969. [DOI] [PubMed] [Google Scholar]
- Garrett S.D. 2nd edn. Pergamon Press; Oxford: 1981. Soil fungi and soil fertility: an introduction to soil mycology. [Google Scholar]
- Geiser D.M., Dorner J.W., Horn B.W. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genetics and Biology. 2000;31:169–179. doi: 10.1006/fgbi.2000.1215. [DOI] [PubMed] [Google Scholar]
- Geiser D.M., Klich M.A., Frisvad J.C. The current status of species recognition and identification in Aspergillus. Studies in Mycology. 2007;59:1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geogianna D.R., Fedorova F.D., Burroughs J.L. Beyond aflatoxin: four distrinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Molecular Plant Pathology. 2010;11:213–226. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons J.G., Rinker D.C. The genomics of microbial domestication in the fermented food environment. Current Opinion in Genetcis & Development. 2015;35:1–8. doi: 10.1016/j.gde.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons J.G., Salichos L., Slot J.C. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Current Biology. 2012;22:1403–1409. doi: 10.1016/j.cub.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M.K., Mack B.M., Wei Q.-J. RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolite gene clusters in Aspergillus flavus. Microbiological Research. 2016;182:150–161. doi: 10.1016/j.micres.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of primer sets for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloer J.B., Rinderknecht B.L., Wicklow D.T. Nominine: a new insecticidal indole diterpene from the sclerotia of Aspergillus nomius. Journal of Organic Chemistry. 1989;54:2530–2532. [Google Scholar]
- Gloer J.B., Tepaske M.R., Sima J.S. Antiinsectan aflavinine derivatives from the sclerotia of Aspergillus flavus. Journal of Organic Chemistry. 1988;53:5457–5460. [Google Scholar]
- Godet M., Munaut F. Molecular strategy for identification in Aspergillus section Flavi. FEMS Microbiology Letters. 2010;304:157–168. doi: 10.1111/j.1574-6968.2009.01890.x. [DOI] [PubMed] [Google Scholar]
- Gonçalves J.S., Ferracin L.M., Viera M.L.C. Molecular analysis of Aspergillus section Flavi isolated from Brazil nuts. World Journal of Microbiology & Biotechnology. 2012;28:1817–1825. doi: 10.1007/s11274-011-0956-3. [DOI] [PubMed] [Google Scholar]
- Gonçalves S., Stchigel A.M., Cano J.P. Aspergillus novoparasiticus: a new clinical species of the section Flavi. Medical Mycology. 2012;50:152–160. doi: 10.3109/13693786.2011.593564. [DOI] [PubMed] [Google Scholar]
- Guezlane-Tebibel N., Bouras N., Mokane S. Aflatoxigenic strains of Aspergillus section Flavi isolated from marketed peanuts (Arachis hypogea) in Algiers (Algeria) Annals of Microbiology. 2013;63:295–305. [Google Scholar]
- Guida V.O. Activitades antibioticas do Aspergillus flavus. Sobre diversas bacterias. Bolletin Societa Paulista Medicale Veterinaria (Sao Paulo) 1948;8:70–73. [Google Scholar]
- Haenni A.L., Robert M., Vetter W. Structure chemique des aspergillomarasmines A and B. Helvetica Chimica Acta. 1965;48:729–750. doi: 10.1002/hlca.19650480409. [DOI] [PubMed] [Google Scholar]
- Hajjaji A., El Otamani M., Bouya D. Occurrence of mycotoxins (ochratoxin A and deoxynivalenol) and toxigenic fungi in Moroccan wheat grains: impact of ecological factors on the growth and ochratoxin A production. Molecular Nutrition and Food Research. 2006;50:494–499. doi: 10.1002/mnfr.200500196. [DOI] [PubMed] [Google Scholar]
- Hamasaki T., Kuwano H., Isono K. New metabolite, parasiticolide A, from Aspergillus parasiticus. Agricultural and Biological Chemistry. 1975;39:749–751. [Google Scholar]
- Hatcher H.J., Schmidt E.L. Nitrification of aspartate by Aspergillus flavus. Applied Microbiology. 1971;21:181–186. doi: 10.1128/am.21.2.181-186.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedayati M.T., Paqualotto A.C., Warn P.A. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology-SGM. 2007;153:1677–1697. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- Hesseltine C.W., Shotwell O.L., Smith M. Production of various aflatoxins by strains of the Aspergillus flavus series. In: Herzberg M., editor. Proceedings of the first joint U.S. – Japan conference on toxic micro-organisms. Mycotoxins. Botulism. UJNR Joint Panels on Toxic Micro-organisms and the U.S. Department of the Interior; Washington D.C., USA: 1970. pp. 202–210. [Google Scholar]
- Hong S.B., Go S.J., Shin H.D., Frisvad J.C., Samson R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97:1316–1329. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- de Hoog G.S., Gerrits van den Ende A.H.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Horn B.W. Aspergillus caelatus, a new species in section Flavi. Mycotaxon. 1997;61:185–191. [Google Scholar]
- Horn B.W., Moore G.G., Carbone I. Sexual reproduction in Aspergillus flavus. Mycologia. 2009;101:423–429. doi: 10.3852/09-011. [DOI] [PubMed] [Google Scholar]
- Horn B.W., Ramirez-Prado J.H., Carbone I. The sexual state of Aspergillus parasiticus. Mycologia. 2009;101:275–280. doi: 10.3852/08-205. [DOI] [PubMed] [Google Scholar]
- Horn B.W., Moore G.G., Carbone I. Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia. 2009;103:174–183. doi: 10.3852/10-115. [DOI] [PubMed] [Google Scholar]
- Horn B.W., Gell R.M., Singh K. Sexual reproduction in Aspergillus flavus sclerotia: Acquisition of novel alleles from soil populations and uniparental mitochrondrial inheritance. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Spierenburg H., Frisvad J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie van Leeuwenhoek. 2012;101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., de Vries R.P., Samson R.A. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Advances in Applied Microbiology. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- Hu X., Xia Q.-W., Zhaom Y.-Y. Speradine B-E, four novel tetracyclic oxindole alkaloids from the marine-derived fungus Aspergillus oryzae. Heterocycles. 2014;89:1662–1669. doi: 10.1248/cpb.c14-00312. [DOI] [PubMed] [Google Scholar]
- Hu X., Xia Q.-W., Zhao Y.-Y. Speradines F-H, three new oxindole alkaloids from the marine-derived fungus Aspergillus oryzae. Chemical and Pharmaceutical Bulletin. 2014;62:942–946. doi: 10.1248/cpb.c14-00312. [DOI] [PubMed] [Google Scholar]
- Hubka V., Kolařík M. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia. 2012;29:1–10. doi: 10.3767/003158512X658123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Lyskova P., Frisvad J.C. Aspergillus pragensis sp. nov. discovered during molecular re-identification of clinical isolates belonging to Aspergillus section Candidi. Medical Mycology. 2014;52:565–576. doi: 10.1093/mmy/myu022. [DOI] [PubMed] [Google Scholar]
- Hubka V., Novakova A., Kolarik M. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani section nov. Mycologia. 2015;107:169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Peterson S.W. A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin producing species. Plants Systematics and Evolution. 2016;302:1267–1299. [Google Scholar]
- Hunter A.J., Jin B., Kelly J.M. Independent duplication of alpha-amylase in different strains of Aspergillus oryzae. Fungal Genetics and Biology. 2011;48:438–444. doi: 10.1016/j.fgb.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Ibarra B.A., Lohmar J.M., Satterlee T. The 14-3-3 protein homolog ArtA regulates developent and secondary metabolism in the opportunistic plant pathogen Aspergillus flavus. Applied and Environmental Microbiology. 2018;84:e02241–e02317. doi: 10.1128/AEM.02241-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka H., Iida M. Maltoryzine, a new toxic metabolite produced by a strain of Aspergillus oryzae var. microsporus isolated from poisonous malt sprout. Nature. 1962;196:681–682. doi: 10.1038/196681a0. [DOI] [PubMed] [Google Scholar]
- Inglis D.O., Binklet J., Skrzypek M.S. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiology. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Peterson S.W., Goto T. Isolation and characterization of Aspergillus nomius from Japanese soil and silkworm excrements. Mycotoxins. 1998;46:9–15. [Google Scholar]
- Ito Y., Peterson S.W., Wicklow D.T. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycological Research. 2001;105:233–239. [Google Scholar]
- Iwasaki T., Kosikowski F.V. Beta-nitropropionic acid in foods. Journal of Food Science. 1973;38:1162–1165. [Google Scholar]
- Jahardhanan K.K., Sattar A., Husain A. Production of fumigaclavine A by Aspergillus tamarii Kita. Canadian Journal of Botany. 1984;30:247–250. doi: 10.1139/m84-036. [DOI] [PubMed] [Google Scholar]
- Junker B., Walker A., Connors N. Production of indole diterpenes by Aspergillus alliaceus. Biotechnology and Bioengineering. 2006;95:919–937. doi: 10.1002/bit.21053. [DOI] [PubMed] [Google Scholar]
- Jurjević Ž., Kubátová A., Kolařík M. Taxonomy of Aspergillus section Petersonii section nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution. 2015;301:2441–2462. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Celiker H., Malikarjunan P.K. Mid-infrared spectroscopy for discrimination and classification of Aspergillus species contamination in peanuts. Food Control. 2015;52:103–111. [Google Scholar]
- Kildgaard S., Mansson M., Dosen I. Accurate dereplication of bioactive secondary metabolites from marine-derived fungi by UHPLC-DAD-QTOFMS and MS/HRMS library. Marine Drugs. 2014;12:3681–3705. doi: 10.3390/md12063681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.Y., Lee J.H., Lee I. An evaluation of aflatoxin and cyclopiazonic acid production in Aspergillus oryzae. Journal of Food Protection. 2014;77:1010–1016. doi: 10.4315/0362-028X.JFP-13-448. [DOI] [PubMed] [Google Scholar]
- Klausmeyer P., McCloud T.G., Tucker K.D. Aspirochlorine class compounds from Aspergillus flavus inhibit azole-resistant Candida albicans. Journal of Natural Products. 2005;68:1300–1302. doi: 10.1021/np050141k. [DOI] [PubMed] [Google Scholar]
- Klich M.A. Aspergillus flavus: the major producer of aflatoxin. Molecular Plant Pathology. 2007;8:713–722. doi: 10.1111/j.1364-3703.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- Klich M.A., Pitt J.I. Differentiation of Aspergillus flavus from Aspergillus parasiticus and other closely related species. Transactions of the British Mycological Society. 1988;91:99–108. [Google Scholar]
- Klitgaard A., Iversen A., Andersen M.R. Aggressive dereplication using UHPLC-DAD-QTOF – screening extracts for up to 3000 fungal secondary metabolites. Analytical and Bioanalytical Chemistry. 2014;406:1933–1943. doi: 10.1007/s00216-013-7582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisel H., Schauer F. VEB Gustav Fischer Verlag; Jena: 1987. Methoden des mykologishen Laboratoriums. [Google Scholar]
- Kretzer A., Li Y., Szaro T. Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia. 1996;88:776–785. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfahl C., Michalka A., Lass-Flörl C. Gliotoxin production by clinical and environmental Aspergillus fumigatus strains. International Journal of Medical Microbiology. 2008;298:319–327. doi: 10.1016/j.ijmm.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Horn B.W., Hesseltine C.W. Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie van Leeuwenhoek. 1987;53:147–158. doi: 10.1007/BF00393843. [DOI] [PubMed] [Google Scholar]
- Laakso J.A., Narske E.D., Gloer J.B. Isokotanins A-C: new bicoumarins from the sclerotia of Aspergillus alliaceus. Journal of Natural Products. 1994;57:128–133. doi: 10.1021/np50103a018. [DOI] [PubMed] [Google Scholar]
- Lan W.J., Wang K.T., Xu M.Y. Secondary metabolites with chemical diversity from the marine-derived fungus Pseudallescheria boydii F19-1 and their cytotoxic activity. RCS Advances. 2016;6:76206–76213. [Google Scholar]
- Lewis R.E., Wiederhold N.P., Lionakis M.S. Frequency and species distribution of gliotoxin-producing Aspergillus isolates recovered from patients in a tertiary-care cancer center. Journal of Clinical Microbiology. 2005;43:6120–6122. doi: 10.1128/JCM.43.12.6120-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesch J.M., Hensens O.D., Springer J.D. Asperlicin, a novel non-peptide cholecystokinin antagonist from Aspergillus alliaceus. Structure elucidation. Journal of Antibiotics. 1985;38:1638–1641. doi: 10.7164/antibiotics.38.1638. [DOI] [PubMed] [Google Scholar]
- Liesch J.M., Hensens O.D., Zink D.L. Novel cholecystokinin antagonists from Aspergillus alliaceus. Journal of Antibiotics. 1988;41:878–881. doi: 10.7164/antibiotics.41.878. [DOI] [PubMed] [Google Scholar]
- Linz J.E., Wee J., Roze L.V. Aspergillus parasiticus SU-1 genome sequence, predicted chromosome structure, and comparative gene expression under aflatoxin-inducing conditions: Evidence that differential expression contributed to species phenotype. Eukaryotic Cell. 2014;13:1113–1123. doi: 10.1128/EC.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Bao L., Wang L. Asperorydines A-M: Prenylated tryptophan-derived alkaloids with neurotrophic effects from Aspergillus oryzae. Journal of Organic Chemistry. 2018;83:812–822. doi: 10.1021/acs.joc.7b02802. [DOI] [PubMed] [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Luo J., Vogel R.F., Niessen L. Rapid deetction of aflatoxin producing fungi in food by real-time quantitative loop-mediated isothermal amplification. Food Microbiology. 2014;44:142–148. doi: 10.1016/j.fm.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Luo J., Taniwaki M.H., Iamanaka B.T. Application of loop-mediated isothermal amplification assays for direct identification of pure cultures of Aspergillus flavus, A. nomius and A. caelatus and for rapid detection in shelled Brazil nuts. International Journal of Food Microbiology. 2014;159:214–224. doi: 10.1016/j.ijfoodmicro.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Luk K.C., Kobbe B., Townsend J.M. Production of cyclopiazonic acid by Aspergillus flavus Link. Applied and Environmental Microbiology. 1977;33:211–212. doi: 10.1128/aem.33.1.211-212.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Peng J., Wu G. Speradines B-D, oxygenated cyclopiazonic aicd alkaloids from the sponge-derived fungus Aspergillus flavus MXH-X104. Tetrahedron. 2015;71:3522–3527. [Google Scholar]
- Ma Y.-M., Ling X.-A., Zhang H.-C. Cytotoxic and antibiotic cyclic pentapeptide from an endophytic Aspergillus tamarii of Ficus carica. Journal of Agricultural and Food Chemistry. 2016;64:3789–3793. doi: 10.1021/acs.jafc.6b01051. [DOI] [PubMed] [Google Scholar]
- Machida M., Asai K., Sano M. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Malysheva S.V., Arroya-Manzanares N., Cary J.W. Identification of novel metabolites from Aspergillus flavus by high resolution and multiple stage mass spectrometry. Food Additives and Contaminants. 2014;31:111–120. doi: 10.1080/19440049.2013.859743. [DOI] [PubMed] [Google Scholar]
- Manabe M., Tanaka K., Goto T. Producing capability of kojic acid and aflatoxin by koji mold. In: Kurata H., Ueno Y., editors. Toxigenic fungi their toxins and health hazards. Vol. 7. Kodansha; Tokyo: 1984. pp. 4–14. (Developments in Food Science). [Google Scholar]
- Marchall É.J. Sur une espèce nouvelle du genre Aspergillus; A. terricola. Revue Mycologie. 1893;1893:101–103. [Google Scholar]
- Martins L.M., de Souza Sant’Anna A., Fungaro M.H.P. The biodiversity of Aspergillus section Flavi and aflatoxins in the Brazilian peanut production chain. Food Research International. 2017;94:101–107. doi: 10.1016/j.foodres.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Marui J., Ohashi-Kunihiro S., Ando T. Penicillin biosynthesis in Aspergillus oryzae and its overproduction by genetic engineering. Journal of Bioscience and Bioengineering. 2010;110:8–11. doi: 10.1016/j.jbiosc.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Marui J., Yamana N., Ohashi-Kunihiro S. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)(2)Cys(6) transcriptional activator and induced by kojic acid at the transcriptional level. Journal of Bioscience and Bioengineering. 2011;112:40–43. doi: 10.1016/j.jbiosc.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Masclaux F., Guého E., de Hoog G.S., Christen R. Phylogenetic relationship of human-pathogenic Cladosporium (Xylohypha) species. Journal of Medical and Veterinary Mycology. 1995;33:327–338. doi: 10.1080/02681219580000651. [DOI] [PubMed] [Google Scholar]
- Massi F.P., Vieira M.L.C., Sartori D. Brazil nuts are subject to infection with B and G aflatoxin-producing fungus, Aspergillus pseudonomius. International Journal of Food Microbiology. 2014;186:14–21. doi: 10.1016/j.ijfoodmicro.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Matsuura S., Yamamoto M., Keneko Y. The structure of the pteridine glycoside from Aspergillus oryzae. Bulletin of the Chemical Society of Japan. 1972;45:492–495. [Google Scholar]
- McAlpin C.E. An Aspergillus flavus mutant produces stipitate sclerotia and synnemata. Mycologia. 2001;93:552–565. [Google Scholar]
- McAlpin C.E., Vesonder R.F., Xie W. A phytotoxic compound produced by Stilbothamnium togoense. Phytopathology. 2000;90:S50. [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. Koeltz Scientific Books; Königstein: 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code): Adopted by the Eighteenth International Botanical Congress, Melbourne, Australia, July, 2011. Regnum Vegetabile 154. [Google Scholar]
- Monti F., Ripamonti F., Hawser S.P. Aspirochlorine: A highly selective and potent inhibitor of fungal protein synthesis. Journal of Antibiotics. 1999;52:311–318. doi: 10.7164/antibiotics.52.311. [DOI] [PubMed] [Google Scholar]
- Moore G.G., Mach G.M., Beltz S.B. Genomic sequence of the aflatoxigenic filamentous fungus Aspergillus nomius. BMC Genomics. 2015;16:551. doi: 10.1186/s12864-015-1719-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G.G., Mack B., Beltz S.B. Draft genome sequence of an aflatoxigenic Aspergillus species, A. bombycis. Genome Biology and Evolution. 2016;8:3297–3300. doi: 10.1093/gbe/evw238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G.G., Mack B.M., Beltz S.B. Genome sequence of an aflatoxigenic pathogen of Argentinean peanut, Aspergillus arachidicola. BMC Genomics. 2018;19:189. doi: 10.1186/s12864-018-4576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton H.E., Kocholaty W., Junowicz-Kocholaty R. Toxicity and antibiotic activity of kojic acid produced by Aspergillus luteo-virescens. Journal of Bacteriology. 1945;50:579–584. [PMC free article] [PubMed] [Google Scholar]
- Mutegi C.K., Nguyi H.K., Hendriks S.L. Factors associated with the incidence of Aspergillus section Flavi and aflatoxin contamination of peanuts in the Busia and Homa Bay districts of western Kenya. Plant Pathology. 2012;61:1143–1153. [Google Scholar]
- Nakamura S., Shimoda Y. Studies on an antibiotic substance oryzacidin, produced by Aspergillus oryzae. V. Existence of β-nitropropionic acid. Journal of the Agricultural Chemical Society of Japan. 1954;28:909–913. [Google Scholar]
- Nesbitt B.F., O'Kelly J., Sargeant K. Toxic metabolites of Aspergillus flavus. Nature. 1962;195:1062–1063. doi: 10.1038/1951062a0. [DOI] [PubMed] [Google Scholar]
- Nielsen K.F., Månsson M., Rank C. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- Nielsen M.L., Nielsen J.B., Rank C. A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiology Letters. 2011;321:157–166. doi: 10.1111/j.1574-6968.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- Nielsen K.F., Smedsgaard J. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardized liquid chromatography-UV-mass spectrometry methodology. Journal of Chromatography A. 2003;1002:111–136. doi: 10.1016/s0021-9673(03)00490-4. [DOI] [PubMed] [Google Scholar]
- Nierman W.C., Yu J., Fedorova-Abrams N.D. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announcements. 2015;3:e00168–e00215. doi: 10.1128/genomeA.00168-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoka N., Assai Y., Nishio M. TMC-2A, -2B, -2C, novel dipeptidyl peptidase IV inhibitors produced by Aspergillus oryzae A374. 1. Taxonomy of producing strain, fermentation and biochemical properties. Journal of Antibiotics. 1977;50:646–652. doi: 10.7164/antibiotics.50.646. [DOI] [PubMed] [Google Scholar]
- Nováková A., Pižl V. Mycoflora in the intestine of Eisenia andrei (Oligochaeta, Lumbricidae) and in vermiculture substrates. Czech Mycology. 2003;55:83–102. [Google Scholar]
- Nozawa K., Nakajima S., Kawai K. Bicoumarins from ascostromata of Petromyces alliaceus. Phytochemistry. 1994;35:1049–1051. [Google Scholar]
- O'Donnell K. Fusarium and its near relatives. In: Reynolds D.R., Taylor J.W., editors. The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. CAB International; Wallingford: 1993. pp. 225–233. [Google Scholar]
- Ohara I. Classification of Aspergillus tamarii-oryzae group. Part 3. Diagnosis of the series, species and subspecies. Research Bulletin Gifu Imperial College of Agriculture. 1953;28:75–85. [Google Scholar]
- Okoth S., Nyongesa B., Ayugi V. Toxigenic potential of Aspergillus species occurring on maize kernels from two agro-ecological zones in Kenya. Toxins. 2012;4:991–1007. doi: 10.3390/toxins4110991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoth S., De Boevre M., Vidal A. Genetic and toxigenic variability within Aspergillus flavus population isolated from maize in two diverse environments in Kenya. Frontiers in Microbiology. 2018;9:57. doi: 10.3389/fmicb.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte R.A., Horn B.W., Dorner J.W. Effect of sexual recombination on population diversity in aflatoxin production by Aspergillus flavus and evidence for cryptic heterokaryosis. Molecular Ecology. 2012;21:1453–1476. doi: 10.1111/j.1365-294X.2011.05398.x. [DOI] [PubMed] [Google Scholar]
- Olarte R.A., Worthington C.J., Horn B.W. Enhanced diversity and aflatoxigenicity in interspecific hydrids of Aspergillus flavus and Aspergillus parasiticus. Molecular Ecology. 2015;24:1889–1909. doi: 10.1111/mec.13153. [DOI] [PubMed] [Google Scholar]
- Olsen M., Johansson P., Möller T. Aspergillus nomius, an important aflatoxin producing species in Brazil nuts? World Mycotoxin Journal. 2008;1:123–126. [Google Scholar]
- Orth R. Mycotoxins of Aspergillus oryzae strains for use in food industry as starters and enzyme-producing molds. Annals de Nutrition et Alimentation. 1977;31:617–624. [PubMed] [Google Scholar]
- Palumbo J.D., O'Keeffe T.L., Mahoney N.E. Inhibition of ochratoxin A production and growth of Aspergillus species by phenolic antioxidant compounds. Mycopathologia. 2007;164:241–248. doi: 10.1007/s11046-007-9057-0. [DOI] [PubMed] [Google Scholar]
- Page R.D.M. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Payne G., Nierman W.C., Wortman J.R. Whole genome comparison of Aspergillus flavus and Aspergillus oryzae. Medical Mycology. 2006;44:S9–S11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- Perrone G., Gallo A., Logrieco A.F. Biodiversity of Aspergillus section Flavi in Europe in relation to management of aflatoxin risk. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone G., Haidukowski M., Stea G. Population structure and aflatoxin production by Aspergillus section from maize in Nigeria and Ghana. Food Microbiology. 2014;41:52–59. doi: 10.1016/j.fm.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Peterson S.W., Ito Y., Horn B.W. Aspergillus bombycis, a new aflatoxigenic species and genetic variation in its sibling species, A. nomius. Mycologia. 2001;93:689–703. [Google Scholar]
- Pildain M.B., Frisvad J.C., Vaamonde G. Two new aflatoxin producing Aspergillus species from Argentinean peanuts. International Journal of Systematic and Evolutionary Microbiology. 2008;58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- Pfefferle W., Anke H., Bross M. Asperfuran, a novel antifungal metabolite from Aspergillus oryzae. Journal of Antibiotics. 1990;43:648–654. doi: 10.7164/antibiotics.43.648. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Hocking A.D., Glenn D.R. An improved medium for the detection of Aspergillus flavus and Aspergillus parasiticus. Journal of Applied Bacteriology. 1983;54:109–114. doi: 10.1111/j.1365-2672.1983.tb01307.x. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Lange L., Lacey A.E. Aspergillus hancockii sp. nov., a biosynthetically talented fungus endemic to southeastern Australian soils. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst C., Njapau H., Cotty P.J. Outbreak of an acute aflatoxicosis in Kenya in 2004: identification of the causal agent. Applied and Environmental Microbiology. 2007;73:2762–2764. doi: 10.1128/AEM.02370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst C., Bandyopadhyay R., Cotty P.J. Diversity of aflatoxin-producing fungi and their impact on food safety in sub-Saharan Africa. International Journal of Food Microbiology. 2014;174:113–122. doi: 10.1016/j.ijfoodmicro.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Probst C., Callicot K.A., Cotty P.J. Deadly strains of Kenyan Aspergillus are distinct from other aflatoxin producers. European Journal of Plant Pathology. 2012;132:419–429. [Google Scholar]
- Probst C., Schulthess F., Cotty P.J. Impact of Aspergillus section Flavi community structure on the development of lethal levels of aflatoxins in Kenyan maize (Zea mays) Journal of Applied Microbiology. 2010;108:600–610. doi: 10.1111/j.1365-2672.2009.04458.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J. 2009. Tracer v. 1.5.http://tree.bio.ed.ac.uk/software/tracer/ Available from: [Google Scholar]
- Rank C., Klejnstrup M.L., Petersen L.M. Comparative chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357) Metabolites. 2012;2:39–56. doi: 10.3390/metabo2010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank C., Nielsen K.F., Larsen T.O. Distribution of sterigmatocystin in filamentous fungi. Fungal Biology. 2011;115:406–420. doi: 10.1016/j.funbio.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Raper K.B., Fennell D.I. Williams & Wilkins; Baltimore: 1965. The genus Aspergillus. [Google Scholar]
- Rayner R.W. CMI and British Mycological Society; Kew, Surrey, England: 1970. A mycological colour chart. [Google Scholar]
- Reategui R., Rhea J., Adophsen J. Leporizine A-C: Epithiodiketopiperazines isolated from Aspergillus species. Journal of Natural Products. 2013;76:1523–1527. doi: 10.1021/np300894y. [DOI] [PubMed] [Google Scholar]
- Riba A., Mokranes S., Mathiu F. Mycoflora and ochratoxin A producing strains of Aspergillus in Algerian wheat. International Journal of Food Microbiology. 2008;122:85–92. doi: 10.1016/j.ijfoodmicro.2007.11.057. [DOI] [PubMed] [Google Scholar]
- Robert M., Barbier M., Lederer E. Two new natural phytotoxins. Aspergillomarasmines A and B and their identity to lycomarasmine and its derivatives. Bulletin de la Societe Chimique de France. 1962;1962:187–198. [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runa F., Carbone I., Bhatnagar D. Nuclear heterogeneity in conidial populations of Aspergillus flavus. Fungal Genetics and Biology. 2015;84:62–72. doi: 10.1016/j.fgb.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Saito M., Tsuruta O. A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. Proceedings of the Japanese Association of Mycotoxicology. 1993;37:31–36. [Google Scholar]
- Saito T. An antibiotic substance produced by Aspergillus oryzae. Shokuryô no Kagaku (Science of Foods) 1946–1947;14 299–300, 326. [Google Scholar]
- Sakata K., Kuwatsuka T., Sakurai A. Isolation of aspirochlorin (= antibiotic A30641) as a true anti-microbial constituent of the antibiotic oryzachlorin, from Aspergillus oryzae. Agricultural and Biological Chemistry. 1983;47:2673–2674. [Google Scholar]
- Sakata K., Maruyama M., Uzawa J. Structural revision of aspirochlorine (=antibiotic A30461), a novel epidithiopiperazine-2,5-dione produced by Aspergillus spp. Tetrahedron Letters. 1987;28:5607–5610. [Google Scholar]
- Sakata K., Masago H., Sakurai A. Isolation of aspirochlorine (= antibiotic A30461) possessing a novel diketopiperazine structure from Aspergillus flavus. Tetrahedron Letters. 1982;23:2095–2098. [Google Scholar]
- Saldan N.C., Almeida R.T.R., Avíncola A. Development of an analytical method for identification of Aspergillus flavus based on chemical markers using HPLC-MS. Food Chemistry. 2018;241:113–121. doi: 10.1016/j.foodchem.2017.08.065. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Gams W. Typification of the species of Aspergillus and associated teleomorphs. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus Systematics. Plenum Press; New York: 1985. pp. 143–154. [Google Scholar]
- Samson R.A., Hoekstra E.S., Frisvad J.C., Filtenborg O. Methods for the detection and isolation of food-borne fungi. In: Samson R.A., Hoekstra E.S., Frisvad J.C., Filtenborg O., editors. Introduction to foodborne fungi. Centraalbureau voor Schimmelcultures; Utrecht (The Netherlands): 1995. pp. 235–242. [Google Scholar]
- Samson R.A., Hong S.-B., Frisvad J.C. Old and new concepts of species differentiation in Aspergillus. Medical Mycology. 2006;44:S133–S144. doi: 10.1080/13693780600913224. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Samson R.A., Visagie C.M. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Seifert K.A. The ascomycete genus Penicilliopsis and its anamorphs. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus systematics. Plenum Press; New York: 1986. pp. 397–428. [Google Scholar]
- Sato N., Horiuchi T., Hamano M. Kojistatin A, a new cysteine protease inhibitor produced by Aspergillus oryzae. Bioscience, Biotechnology and Biochemistry. 1996;60:1747–1748. [Google Scholar]
- Sato A., Oshima K., Noguchi H. Draft genome sequencing and comparative analysis of Aspergillus sojae NBRC 34239. DNA Research. 2011;18:165–176. doi: 10.1093/dnares/dsr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H.W. Effect of corn steep liquor on mycelial growth and aflatoxin production in Aspergillus parasiticus. Applied Microbiology. 1966;14:381–385. doi: 10.1128/am.14.3.381-385.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y., Takahashi S., Osada H. Identification of a novel sesquiterpene biosynthetic machinery involved in astellolide biosynthesis. Scientific Reports. 2016;6 doi: 10.1038/srep32865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y., Kawatani M., Futamura Y. An overproduction of astellolides induced by genetic disruption of chromatin-remodelling factors in Aspergillus oryzae. Journal of Antibiotics. 2016;69:4–8. doi: 10.1038/ja.2015.73. [DOI] [PubMed] [Google Scholar]
- Shiomi K., Hatae K., Yamaguchi Y. New antibiotics miyakamides produced by a fungus. Journal of Antibiotics. 2002;55:952–961. doi: 10.7164/antibiotics.55.952. [DOI] [PubMed] [Google Scholar]
- Smedsgaard J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. Journal of Chromatography A. 1997;760:264–270. doi: 10.1016/s0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Alachiotis N. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics. 2010;26:i132–i139. doi: 10.1093/bioinformatics/btq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares C., Rodriguez P., Peterson S.W. Three new species of Aspergillus section Flavi isolated from almonds and maize in Portugal. Mycologia. 2012;104:682–697. doi: 10.3852/11-088. [DOI] [PubMed] [Google Scholar]
- Sobolev V.S., Cole R.J., Dorner J.W. Isolation and structure elucidation of a new metabolite produced by Aspergillus parasiticus. Journal of Natural Products. 1997;60:847–850. [Google Scholar]
- Son B.W., Choi J.S., Kim J.C. Parasitenone, a new epoxycyclohexenone related to gabosine from the marine-derived fungus Aspergillus parasiticus. Journal of Natural Products. 2002;65:794–795. doi: 10.1021/np010450k. [DOI] [PubMed] [Google Scholar]
- Springer J.P., Büchi G., Kobbe B. The structure of ditryptophenaline – a new metabolite of Aspergillus flavus. Tetrahedron Letters. 1977;18:2403–2406. [Google Scholar]
- States J.S., Christensen M. Aspergillus leporis a new species related to Aspergillus flavus. Mycologia. 1966;58:738–742. [Google Scholar]
- Staub G.M., Gloer J.B., Wicklow D.T. Aspernomine: a new cytotoxic antiinsectan metabolite with a novel ring system from the sclerotia of Aspergillus nomius. Journal of the American Chemical Society. 1992;114:1015–1017. [Google Scholar]
- Staub G.M., Gloer K.B., Gloer J.B. New paspalinine derivatives from the sclerotia of Aspergillus nomius. Tetrahedron Letters. 1993;34:2569–2572. [Google Scholar]
- Stierle A.A., Stierle D.B., Bugni T. Sequoiatones A and B: Novel antitumor metabolites isolated from a redwood endophyte. Journal of Organic Chemistry. 1999;64:5479–5484. doi: 10.1021/jo990277l. [DOI] [PubMed] [Google Scholar]
- Stierle A.A., Stierle D.B., Bugni T. Sequioatones C-F, constituents of the redwood endophyte Aspergillus parasiticus. Journal of Natural Products. 2001;64:1350–1353. doi: 10.1021/np010022e. [DOI] [PubMed] [Google Scholar]
- Stierle D.B., Stierle A.A., Bugni T. Sequioamonascins A-D: novel anticancer metabolites isolated from a redwood endophyte. Journal of Organic Chemistry. 2003;68:4966–4969. doi: 10.1021/jo0340253. [DOI] [PubMed] [Google Scholar]
- Stubblefield R.D., Shotwell O.L., Shannon G.M. Parasiticol – a new metabolite from Aspergillus parasiticus. Journal of Agricultural and Food Chemistry. 1970;18:391–393. doi: 10.1021/jf60169a025. [DOI] [PubMed] [Google Scholar]
- Sun K., Li Y., Guo L. Indole diterpenoids and isocoumarin from the fungus, Aspergillus flavus, isolated from the prawn Penaeus vannamei. Marine Drugs. 2014;12:3970–3981. doi: 10.3390/md12073970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamogami S., Katayama M., Marumo S. Synthesis of 5-demethyl-6-deoxy analogue of sporogen AO1, a sporogenic substance produced by Aspergillus oryzae. Bioscience, Biotechnology and Biochemistry. 1996;60:1372–1374. [Google Scholar]
- Tanaka K., Goto T., Manabe M. Traditional Japanese fermented food free from mycotoxin contamination. JARQ – Japan Agricultural Research Quarterly. 2002;36:45–50. [Google Scholar]
- Tang M.C., Lin H.C., Li D.H. Discovery of unclustered fungal indole diterpene biosynthetic pathways through combinatorial pathway reassembly in engineered yeast. Journal of the American Chemical Society. 2015;137:13724–13727. doi: 10.1021/jacs.5b06108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniwaki M.H., Frisvad J.C., Ferranti L.S. Biodiversity of mycobiota throughout the Brazil nut supply chain: From rainforest to consumer. Food Microbiology. 2017;61:14–22. doi: 10.1016/j.fm.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Taniwaki M.H., Pitt J.I., Iamanaka B.T. Aspergillus bertholletius sp. nov. from Brazil nuts. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.W., Jacobson D.J., Kroken S. Phylogenetic species recognition and species concepts in Fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- TePaske M.R., Gloer J.B., Wicklow D.T. Aflavazole – a new intiinsectan carbazole metabolite from the sclerotia of Aspergillus flavus. Journal of Organic Chemistry. 1990;55:5299–5301. [Google Scholar]
- TePaske M.R., Gloer J.B., Wicklow D.T. Leporin A – an antiinsectan N-alkoxypyridone from the sclerotia of Aspergillus leporis. Tetrahedron Letters. 1991;32:5687–5690. [Google Scholar]
- TePaske M.R., Gloer J.B., Wicklow D.T. Aflavarin and beta-aflatrem – new antiinsectan metabolites from the sclerotia of Aspergillus flavus. Journal of Natural Products. 1992;55:1080–1086. doi: 10.1021/np50085a013. [DOI] [PubMed] [Google Scholar]
- Terebayashi Y., Sano M., Yamani N. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genetics and Biology. 2010;47:953–961. doi: 10.1016/j.fgb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Thom C., Church M.B. Williams and Wilkins; Baltimore: 1926. The Aspergilli. [Google Scholar]
- Thom C., Raper K.B. Williams and Wilkins; Baltimore: 1945. A manual of the Aspergilli. [Google Scholar]
- Tokuoka M., Kikuchi T., Shinohara Y. Cyclopiazonic acid biosynthetic cluster gene cpaM is required for speradine A biosynthesis. Bioscience, Biotechnology and Biochemistry. 2015;79:2081–2085. doi: 10.1080/09168451.2015.1065167. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Mugishima T., Komatsu K. Speradine A, a new pentacyclic oxindole alkaloid from a marine-derived fungus Aspergillus tamarii. Tetrahedron. 2003;59:3227–3230. [Google Scholar]
- Turner W.B. Academic Press; London: 1971. Fungal metabolites. [Google Scholar]
- Turner W.B., Aldridge D.C. Academic Press; London: 1983. Fungal metabolites II. [Google Scholar]
- Uka V., Moore G.G., Arroyo-Manzanares N. Unravelling the diversity of the cyclopiazonic acid family of mycotoxins in Aspergillus flavus by UHPLC triple-TOF HRMS. Toxins. 2017;9:35. doi: 10.3390/toxins9010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura M., Koyama Y., Takeda I. Fine de novo sequencing of a fungal genome using only SOLiD short read data: verification on Aspergillus oryzae RIB 40. PloS One. 2013;8 doi: 10.1371/journal.pone.0063673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura M., Koike H., Nagano T. MIDDAS-M: Motif-independent de novo detection of secondary metabolite gene clusters through the integration og genome sequencing and transcriptome data. PloS One. 2013;8 doi: 10.1371/journal.pone.0084028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura M., Nagano N., Koike H. Characterization of the biosynthetic gene cluster for the ribosomally sunthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genetics and Biology. 2014;68:23–30. doi: 10.1016/j.fgb.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Van der Merwe K.J., Steyn P.S., Fourie L. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature. 1965;205:1112–1113. doi: 10.1038/2051112a0. [DOI] [PubMed] [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. A reappraisal of fungi producing aflatoxin. World Mycotoxin Journal. 2009;2:263–277. [Google Scholar]
- Varga J., Baranyi N., Chandrasekaran M. Mycotoxin production in the genus Aspergillus: an update. Acta Biologica Szegediensis. 2015;59:151–167. [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. Two new aflatoxin producing species and an overview of Aspergillus section Flavi. Studies in Mycology. 2011;69:57–80. doi: 10.3114/sim.2011.69.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Kevei E., Palagyi A. Genetic variability within the toxigenic Petromyces genus. Cereal Research Communications. 1997;25:285–289. [Google Scholar]
- Viaro H.P., da Silva J.J., Ferranti L.D. The first report of Aspergillus novoparasiticus, A. arachidicola and A. pseudocaelatus in Brazilian corn kernels. International Journal of Food Microbiology. 2017;243:46–51. doi: 10.1016/j.ijfoodmicro.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Wagacha J.M., Mutegi C., Karanja Fungal species isolated from peanuts in major Kenyan marketed peanuts: Emphasis on Aspergillus section Flavi. Crop Protection. 2013;52:1–9. [Google Scholar]
- Walker J.C., Murphy A. Onion-bulb decay caused by Aspergillus alliaceus. Phytopathology. 1934;24:289–291. [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., editor. PCR protocols: a guide to methods and applications. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]
- White E.C., Hill J.H. Studies in the antibacterial products formed by moulds. I. Aspergillic acid, a product of a strain of Aspergillus flavus. Journal of Bacteriology. 1943;45:433–443. doi: 10.1128/jb.45.5.433-443.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow D.T. Adaptation in wild and domesticated yellow-green aspergilli. In: Kurata H., Ueno Y., editors. Toxigenic fungi their toxins and health hazards. Vol. 7. Kodansha; Tokyo: 1984. pp. 78–86. (Developments in Food Science). [Google Scholar]
- Wicklow D.T. Aspergillus leporis sclerotium formation on rabbit dung. Mycologia. 1985;77:531–534. [Google Scholar]
- Wicklow D.T., Dowd P.F., Alfatafta A.A. Ochratoxin A: An antiinsectan metabolite from the sclerotia of Aspergillus carbonarius NRRL 369. Canadian Journal of Microbiology. 1996;42:1100–1103. doi: 10.1139/m96-141. [DOI] [PubMed] [Google Scholar]
- Wicklow D.T., McAlpin C.E. Cultural conditions promoting sclerotium formation in Stilbothamnium togoense. Mycologia. 1990;82:165–169. [Google Scholar]
- Wicklow D.T., Shotwell O.L. Intrafungal distribution of aflatoxins among conidia and sclerotia of Aspergillus flavus and Aspergillus parasiticus. Canadian Journal of Microbiology. 1983;29:1–5. doi: 10.1139/m83-001. [DOI] [PubMed] [Google Scholar]
- Wicklow D.T., McAlpin C.E., Peterson S.W. Common genotypes (RFLP) within a diverse collection of yellow-green aspergilli used to produce traditional Oriental fermented foods. Mycoscience. 2002;43:289–297. [Google Scholar]
- Wicklow D.T., Vesonder R.F., McAlpin C.E. Examination of Stilbothamnium togoense for Aspergillus flavus group mycotoxins. Mycotaxon. 1989;34:249–252. [Google Scholar]
- Yamada T., Hiratake J., Aikawa M. Cysteine protease inhibitors produced by the industrial koji mold, Aspergillus oryzae O-1018. Bioscience, Biotechnology and Biochemistry. 1998;62:907–914. doi: 10.1271/bbb.62.907. [DOI] [PubMed] [Google Scholar]
- Ye Y., Minami A., Igarashi Y. Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin B in filamentous fungi. Angewandte Chemie International Edition. 2016;55:8072–8075. doi: 10.1002/anie.201602611. [DOI] [PubMed] [Google Scholar]
- Yokotsuka T., Oshita K., Kikuchi T. Studies on the compounds produced by molds. Part VI. Aspergillic acid, kojic acid, β-nitropropionic acid and oxalic acid in solid-koji. Journal of the Agricultural Chemical Society. 1969;43:189–196. [Google Scholar]
- Zeringue H.J., Jr., Shin B.Y., Maskos K. Identification of the bright-greenish-yellow-fluorescence (BGY-F) compound on cotton lint associated with aflatoxin contamination in cottonseed. Phytochemistry. 1999;52:1391–1397. doi: 10.1016/s0031-9422(99)00432-x. [DOI] [PubMed] [Google Scholar]
- Zhao G., Yao Y., Chen W. Comparison and analysis of the genomes of two Aspergillus oryzae strains. Journal of Agricultural and Food Chemistry. 2013;61:7805–7809. doi: 10.1021/jf400080g. [DOI] [PubMed] [Google Scholar]
- Zhao G., Yao Y., Hou L. Draft genome sequence of Aspergillus oryzae 100-8, an increased acid protease production strain. Genome Announcements. 2014;2:e00548–e00614. doi: 10.1128/genomeA.00548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Yao Y., Qi W. Draft genome sequence of Aspergillus oryzae strain 3-042. Eukaryotic Cell. 2012;11:1178. doi: 10.1128/EC.00160-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.