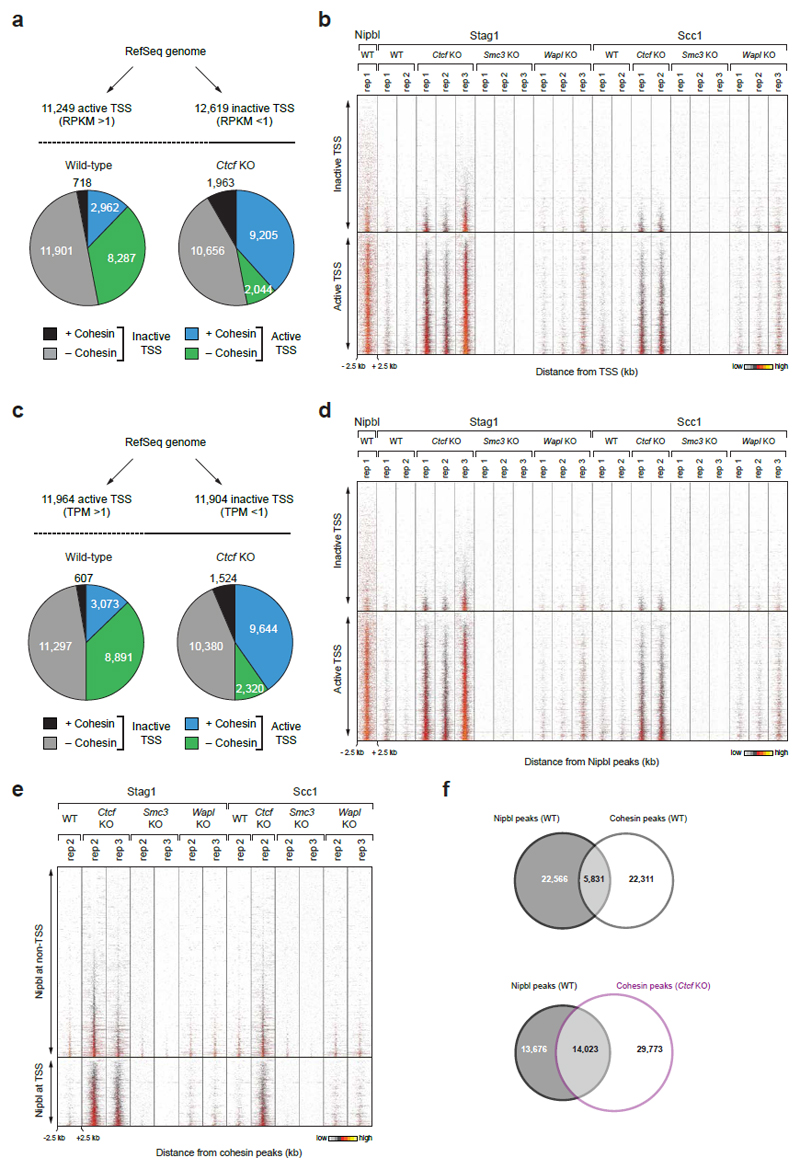
Extended Data Figure 4. Cohesin redistribution to transcriptional start sites in Ctcf KO MEFs.
a, Binding of cohesin at transcription start sites (TSSs) of active and inactive genes, which were defined by RNA-seq analysis. Genes with an RPKM > 1 were considered as active, whereas genes with an RPKM < 1 were classified as inactive. Pie charts indicate the relative binding of cohesin at all annotated TSSs of the RefSeq genome (mm9) in WT and Ctcf KO MEFs. b, Density heat map of cohesin and Nipbl binding at active and inactive TSSs as defined in a. Active and inactive TSSs were sorted according to the read density of Stag1 binding in Ctcf KO cells (replica 3). c, Binding of cohesin at TSSs of active and inactive genes, which were defined by GRO-seq analysis. Genes with a TPM > 1 were considered as active, whereas genes with an TPM < 1 were classified as inactive. Pie charts indicate the relative binding of cohesin at all annotated TSSs of the RefSeq genome (mm9) in WT and Ctcf KO cells. d, Density heat map of cohesin and Nipbl binding at active and inactive TSSs as defined in c. Active and inactive TSSs were sorted according to the read density of Stag1 binding in Ctcf KO (replica 3). A density scale from low (grey) to high (yellow) is shown (b,d). e, Heat map of Nipbl and cohesin binding at Nipbl peaks in MEFs of the indicated genotypes. The Nipbl peaks were subdivided according to their TSS localization. Peaks were sorted according to the Stag1 binding density in Ctcf KO cells (replica 3). f, Venn diagram indicating the overlap between Nipbl and cohesin peaks in WT or Ctcf KO MEFs.