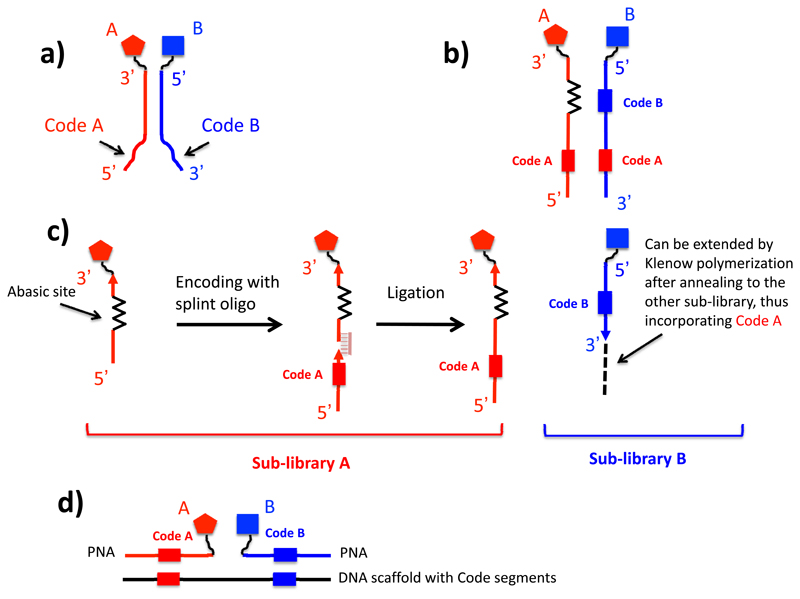
Figure 6.
Encoding strategies for dual-pharmacophore encoded chemical libraries. (a) General structure of ESAC library members, which are decoded by hybridization onto oligonucleotide microarrays. (b) In a more powerful procedure, ESAC library members are encoded with a methodology, that leads to the simultaneous presence of two codes (identifying building blocks A and B) onto one of the two complementary strands. (c) Encoding strategy, compatible with high-throughput sequencing decoding procedures. Members of the “red” sublibrary are constructed by the coupling of building blocks to a general oligonucleotide carrying an abasic site, followed by encoding by ligation assisted by a splint degradable oligo. By contrast, members of the “blue” sublibrary are created by coupling building blocks to the 5’ extremity of suitable oligonucleotides, carrying an identification code. The two sublibraries can then be annealed followed by a Klenow polymerization step. (c) In one of the possible implementations of the technology, chemically-modified PNAs are hybridized to DNA templates, which carry suitable complementary coding sequences.