Abstract
Acephalic spermatozoa syndrome is a severe teratozoospermia that leads to male infertility. Our previous work showed that biallelic SUN5 mutations are responsible for acephalic spermatozoa syndrome in about half of affected individuals, while pathogenic mechanisms in the other individuals remain to be elucidated. Here, we identified a homozygous nonsense mutation in the testis-specific gene PMFBP1 using whole-exome sequencing in a consanguineous family with two infertile brothers with acephalic spermatozoa syndrome. Sanger sequencing of PMFBP1 in ten additional infertile men with acephalic spermatozoa syndrome and without SUN5 mutations revealed two homozygous variants and one compound heterozygous variant. The disruption of Pmfbp1 in male mice led to infertility due to the production of acephalic spermatozoa and the disruption of PMFBP1’s cooperation with SUN5 and SPATA6, which plays a role in connecting sperm head to the tail. PMFBP1 mutation-associated male infertility could be successfully overcome by intracytoplasmic sperm injection (ICSI) in both mouse and human. Thus, mutations in PMFBP1 are an important cause of infertility in men with acephalic spermatozoa syndrome.
Keywords: male infertility, acephalic spermatozoa syndrome, PMFBP1, head-tail coupling apparatus, ICSI
Introduction
Infertility affects approximately 15% of couples worldwide, and nearly half of the infertility cases are attributed to the male.1 Numerous cases of male infertility are caused by morphological defects of the spermatozoa, which decrease or abolish the fertilization and is termed teratozoospermia.2 Acephalic spermatozoa syndrome (SPGF16 [MIM: 617187]) is a rare but severe type of teratozoospermia that is characterized by the predominance of headless spermatozoa in the ejaculate.3, 4 In some individuals with acephalic spermatozoa, the syndrome has been identified to be familial, strongly suggesting that this syndrome has a genetic origin.5, 6, 7, 8, 9 Many male infertility mouse models with decapitated or acephalic spermatozoa have been reported,10, 11, 12, 13, 14, 15, 16, 17 but no fertility-associated mutations found in these mice have been detected in infertile men with acephalic spermatozoa. Our recent work found that biallelic SUN5 (Sad1 and UNC84 domain containing 5, also known as SPAG4L [HGNC:16252, MIM: 613942]) mutations are responsible for autosomal-recessive acephalic spermatozoa syndrome in 47.06% of affected individuals in our cohort and that the ablation of Sun5 leads to acephalic spermatozoa in a mouse model, indicating that defects in SUN5 may be a main cause of the acephalic spermatozoa syndrome.18, 19 Nevertheless, SUN5 mutations explained only about half of the infertile men with acephalic spermatozoa, and the pathogenic mechanisms in the genetically unexplained acephalic spermatozoa syndrome remain to be elucidated.
Here, we identified a homozygous mutation in PMFBP1 (polyamine modulated factor 1 binding protein 1 [HGNC:17728, GenBank: NM_031293.2]) gene using whole-exome sequencing (WES) in a consanguineous family with two infertile brothers with acephalic spermatozoa syndrome. Subsequent Sanger sequencing of ten unrelated infertile men with acephalic spermatozoa syndrome and without SUN5 mutations detected two new homozygous nonsense mutations and one compound mutation. These results suggest that the PMFBP1 mutations, in addition to SUN5 mutations, are also responsible for acephalic spermatozoa syndrome. Using CRISPR/Cas9 technology, we generated Pmfbp1 knockout mice and found that the disruption of Pmfbp1 led to male infertility due to the production of acephalic spermatozoa. PMFBP1 is localized at the head-tail coupling apparatus (HTCA) and cooperates with SUN5 and SPATA6 to connect sperm head to tail. PMFBP1 mutation-associated infertility could be successfully overcome by intracytoplasmic sperm injection (ICSI) in both mouse and human. Thus, mutations in PMFBP1 are another major cause of acephalic spermatozoa syndrome.
Material and Methods
Study Participants
The study cohort was composed of 16 infertile men from the Reproductive Medicine Center, Department of Obstetrics and Gynecology at the First Affiliated Hospital of Anhui Medical University in Hefei, China and 7 men from the Center of Clinical Reproductive Medicine, First Affiliated Hospital, Nanjing Medical University in Nanjing, China. Apart from the 17 men described in our previous study,18 6 additional men, including 2 brothers from a consanguineous family and 4 sporadic affected individuals, were recruited in this study. The diagnosis in infertile men with acephalic spermatozoa syndrome was at least twice confirmed by means of semen analysis and Papanicolaou staining performed according to the guidelines of the World Health Organization. All infertile men with acephalic spermatozoa syndrome had normal karyotype (46, XY) and negative results on Y chromosome microdeletion. No testicular biopsies were performed. Three semen samples and 100 DNA samples of unrelated, anonymous, native male donors were used as controls. The study protocol was approved by the ethics committee of Anhui Medical University and informed consent was obtained from all participants.
Genetic Analysis
Genomic DNA was extracted from peripheral blood leukocytes. Whole-exome sequencing was performed for the infertile man of a consanguineous family to identify the causative mutations. The Sure Select Human All Exon V5 (Agilent Technologies, 5190-6208) was used for whole-exome capture and enrichment according to the manufacture’s protocol. Exome sequencing was performed on the HiSeq2000 sequencing platform (Illumina). The Burrows-Wheeler Alignment software was used for aligning sequence reads to the human reference sequence (UCSC Genome Browser hg19). Sequence variants including single-nucleotide polymorphisms and insertion/deletions were annotated by ANNOVAR software.20 Confirmation of mutation of probands and the familial cosegregation analysis were used for Sanger sequencing. Mutation screening for mutations in other unrelated male infertile men with acephalic spermatozoa syndrome was also conducted by direct Sanger sequencing.
The Generation of Pmfbp1 Knockout Mice
The T7 promoter and the guiding sequence were added to the sgRNA by PCR amplification using the following primers: Pmfbp1-S5-F: 5′-ATAGCTTCACTTAACGGGCTGACA-3′; Pmfbp1-S5-R: 5′-AAACTGTCAGCCCGTTAAGTGAAG-3′; Pmfbp1-S8-F: 5′-ATAGACTGCCCCCGAGAGGAC-3′; Pmfbp1-S8-R: 5′-AAACGTCCTCTCGGGGGCAGT-3′. B6D2F1 (C57BL/6 × DBA2, RRID: IMSR_JAX:100006) female mice and ICR female mice were used as embryo donors and foster mothers, respectively. Superovulated female B6D2F1 mice (6 to 8 weeks old) were mated with B6D2F1 stud males, and the fertilized embryos were collected from the oviducts. Cas9 mRNA (20 ng) and sgRNA (10 ng) were injected into the cytoplasm of fertilized eggs with well-recognized pronuclei in M2 medium (Sigma, M7167-50 mL). The injected zygotes were cultured in KSOM (modified simplex-optimized medium, Millipore) with amino acids at 37°C under 5% CO2 in air, and then 15–25 blastocysts were transferred into the uterus of pseudopregnant ICR females. The genotyping primers were as follows: forward, 5′-GATGAGTAATAACAGCCCAGG-3′ and reverse, 5′-CTTGATGCATGCGCAGTTAG-3′ for wild-type allele; forward, 5′-GATCTCAGACTCCCTCGTCAACTC-3′ and reverse, 5′-GGAGTGGGCAGGATTGAAAG-3′ for Pmfbp1 knockout allele. All of the animal experiments were performed according to approved institutional animal care and use committee (IACUC) protocols (#08-133) of the Institute of Zoology, Chinese Academy of Sciences.
Antibodies
Rabbit anti-PMFBP1 polyclonal antibody (HPA008518) was purchased from Sigma-Aldrich. Mouse anti-PMFBP1 polyclonal antibody (ab67871) was purchased from Abcam. Rabbit anti-PMFBP1 polyclonal antibody (orb158179) for immunoblotting was purchased from Biorbyt. Rabbit anti-SUN5 polyclonal antibody (17495-1-AP, RRID: AB_1939754) was purchased from Proteintech. Mouse anti-sp56 antibody (55101, RRID: AB_130101) was purchased from QED Bioscience. Anti-FLAG (M20008), anti-Pan-Actin (M20010L), and anti-GST (M20007L) antibodies were purchased from Abmart. Rabbit anti-SPATA6 polyclonal antibody was a gift from Wei Yan (University of Nevada). Goat anti-rabbit FITC (ZF-0311), goat anti-mouse FITC (ZF-0312), and goat anti-mouse TRITC (ZF-0313)-conjugated secondary antibodies were purchased from Zhong Shan Jin Qiao (Beijing, China). FITC-peanut agglutinin (L7381) was purchased from Sigma-Aldrich. Alexa Fluor 680-conjugated goat anti-mouse (A21057) and Alexa Fluor 680-conjugated goat anti-rabbit (A21109) for immunoblotting were purchased from Invitrogen.
Assessment of the Fertility of Pmfbp1-Deficient Mice
The fertility assessment experiments were performed as previously described.21 Each male mouse (8 or 9 weeks old) was caged with 2 wild-type CD1 females (7 or 8 weeks old), and their vaginal plugs were checked every morning. The plugged female was separated and single caged, and the pregnancy results were recorded. If a female did not generate any pups by day 22 postcoitus, the mouse was deemed not pregnant and euthanized to confirm that result. Each male underwent 6–10 cycles of the above breeding assay.
Epididymal Sperm Count
The caudal epididymis was dissected from 8- or 9-week-old mice. Spermatozoa were squeezed out from the caudal epididymis and incubated for 30 min at 37°C under 5% CO2. The incubated sperm medium was then diluted 1:500 and transferred to a hemocytometer for counting.
Tissue Collection and Histological Analysis
The caudal epididymis from at least three mice for each genotype were dissected immediately after euthanasia, fixed in 4% (mass/vol) paraformaldehyde (PFA; Solarbio, P1110) for up to 24 hr, stored in 70% (vol/vol) ethanol, and embedded in paraffin. The 5 μm sections were prepared and mounted on glass slides. After deparaffinization, slides were stained with H&E for histological analysis. For PAS staining, testes were fixed by perfusing mice with Bouin’s fixatives (Polysciences). After deparaffinization, slides were stained with PAS and hematoxylin. Stages of seminiferous epithelium cycle and spermatid development were determined as previously described.22
Transmission Electron Microscopy Analysis
The adult mouse testes and caudal epididymis were dissected and fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer overnight. After washing in 0.1 M cacodylate buffer, the tissues were cut into small pieces of approximately 1 mm3 and immersed in 1% OsO4 for 1 hr at 4°C. Then, the samples were dehydrated through a graded acetone series and embedded in resin (Low Viscosity Embedding Media Spurr’s Kit, EMS, 14300). Ultrathin sections were cut on an ultramicrotome, stained with uranyl acetate and lead citrate, and observed.
Immunofluorescence
The spermatozoa were spread on glass slides for morphological observation or immunostaining. After air drying, spermatozoa were fixed in 4% PFA at room temperature, and slides were washed with PBS three times and blocked with 5% bovine serum albumin (Amresco, AP0027). The primary antibody was added to the sections and incubated at 4°C overnight, followed by incubation with the secondary antibody. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The IF images were taken immediately using an LSM 780/710 microscope (Zeiss) or SP8 microscope (Leica).
Immunoprecipitation
SUN5 and SPATA6 were cloned into pRK vector and PMFBP1 was cloned into pEGFP-C1 vector. pRK-FLAG-SUN5, pEGFP-Pmfbp1, pRK-FLAG-Spata6 and pEGFP-Pmfbp1 were co-transfected into HEK293T cells. Transfected cells were lysed in TAP lysis buffer (50 mM HEPES-KOH [pH 7.5], 100 mM KCl, 2 mM EDTA, 10% glycerol, 0.1% NP-40 10 mM NaF, 0.25 mM Na3VO4, 50 mM β-glycerolphosphate) plus protease inhibitors (Roche, 04693132001) for 30 min on ice and centrifuged at 13,000 × g for 15 min. For immunoprecipitation, cell lysates were incubated with anti-FLAG antibody overnight at 4°C and then incubated with protein A-Sepharose (GE, 17-1279-03) for 2 hr at 4°C. Thereafter, the precipitants were washed two times with IP buffer (20 mM Tris [pH 7.4], 2 mM EGTA, 1% NP-40), and the immune complexes were eluted with sample buffer containing 1% SDS for 10 min at 55°C and analyzed by immunoblotting.
GST Pull-Down Experiments
GST-SUN5C was expressed as described before.23 PMFBP1 and SPATA6 were cloned into pMAL-c2X vector and expressed in E. coli. GST-SUN5C was incubated with Glutathione Sepharose 4B (GE Healthcare) at 4°C for 2 hr. The GST protein was used as a negative control. The GST beads were centrifuged and washed with a high-salt buffer (50 mM Tris [pH 7.4], 150 mM potassium chloride, 2 mM magnesium chloride, 0.1% Triton). MBP-PMFBP1 or MBP-SPATA6 were added, and the beads were incubated at 4°C for 2 hr. The beads were centrifuged and washed three times with the high-salt buffer, and SDS loading buffer was added. The samples were analyzed by immunoblotting with anti-GST, anti-PMFBP1, or anti-SPATA6 antibodies.
ICSI for Mice
CD1 and B6D2F1 mice (8 to 12 weeks old) were used to prepare mature oocyte donors. Spermatozoa were released from the testes using HTF (human tubal fluid) medium. WT spermatozoa were decapitated by mild sonication. WT and SUN5-null sperm heads were collected by centrifugation in 60% Percoll (Sigma, P4937) followed by three washes in M2 medium. Single sperm heads were picked up from the sperm suspension and injected into WT oocytes using a micromanipulator with a Piezoelectric actuating pipette at room temperature. Injected oocytes were transferred to the KSOM medium under mineral oil and cultured at 37°C in a humidified atmosphere with 5% CO2. The injected oocytes were analyzed 5–8 hr after ICSI and transferred into the oviducts of pseudo-pregnant CD1 females that had been mated during the previous night with vasectomized males. Full-term pups derived from ICSI embryos were obtained through natural labor.
ICSI for PMFBP1 Mutation-Associated Infertile Men
Written consent was obtained from the couples, and they selected to undergo intracytoplasmic sperm injection (ICSI) in the Reproductive Medicine Center, Department of Obstetrics and Gynecology at the First Affiliated Hospital of Anhui Medical University in Hefei, China. Both female partners had undergone a long protocol pituitary downregulation using GnRH agonist (Triptorelin, Diphereline 3.75 mg, Ipsen Pharma Biotech) and control ovarian hyperstimulation by recombinant FSH (Gonal-F; Serono) Oestradiol plasma levels and follicle growth were monitored every 2 days and human chorionic gonadotrophin (HCG, Livzon Pharmaceutical) was administered when three or more than follicles reached 18 mm in diameter. Oocyte retrieval was performed 36 hr after HCG injection. For individuals 1 and 2 (family 1: II-2 and II-3), 6 and 23 oocytes were retrieved, respectively; there were 4 and 19 mature oocytes (MII) for ICSI. For individual 3 (family 3: II-4), 16 oocytes were retrieved 36 hr after HCG injection; 14 oocytes were at the MII stage. Spermatozoa were prepared by discontinuous density gradient centrifugation and the resulting suspension was diluted in 10 μL drops of polyvinyl pyrolidine (PVP) covered with oil. During the ICSI, the immobilization procedure of the abnormal head-tail junction spermatozoa was modified to gentle compression of various regions of the tail because the head-tail junction was deemed fragile. We successfully injected a head-tail sperm into each oocyte. The development of embryos are shown in Table S3. For individuals 1 and 2, we obtained two day-5 blastocysts (4BB, 3BB) for the elder brother and four day-5 blastocysts (4BB, 3BB, 3BB, 3BB) and two day-6 blastocysts (4AA, 3BB) for the younger brother, according to the scoring system of Gardner and Schoolcraft.24 For the elder brother, the two day-5 blastocysts were immediately transferred, and a singleton clinical pregnancy was confirmed and resulted in the delivery of a healthy boy whose birth weight at full-term was 3,000 g. For the younger brother, all embryos were cryopreserved to prevent ovarian hyperstimulation syndrome (OHSS). After 2 months, two day-5 blastocysts and two day-5 blastocysts were thawed and transferred in two successive artificial cycles but resulted in two failed pregnancies. A clinical pregnancy was obtained during the third attempt with two day-6 blastocysts, resulting in the birth of one 3,400 g healthy boy. For individual 3, the GnRH antagonist protocol was used for his wife, 16 oocytes were retrieved, 14 mature oocytes were therefore injected, and we obtained six day-5 blastocysts (4AA, 4BB, 3BB, 3BB, 3BB, 3BB) and three day-6 blastocysts, according to the scoring system of Gardner and Schoolcraft.24 All embryos were cryopreserved. After 2 months, two day-5 blastocysts (4AA, 4BB) were thawed and transferred in the first artificial cycle, resulting in a normal ongoing twin pregnancy. Currently, the ultrasound at 18 weeks looks completely normal.
Statistical Analysis
All data are presented as the mean ± SEM. The statistical significance of the differences between the mean values for the different genotypes was measured by the Student’s t test with a paired, 2-tailed distribution. The data were considered significant when the p value was less than 0.05 (∗) or 0.01 (∗∗).
Results
PMFBP1 and Acephalic Spermatozoa Syndrome
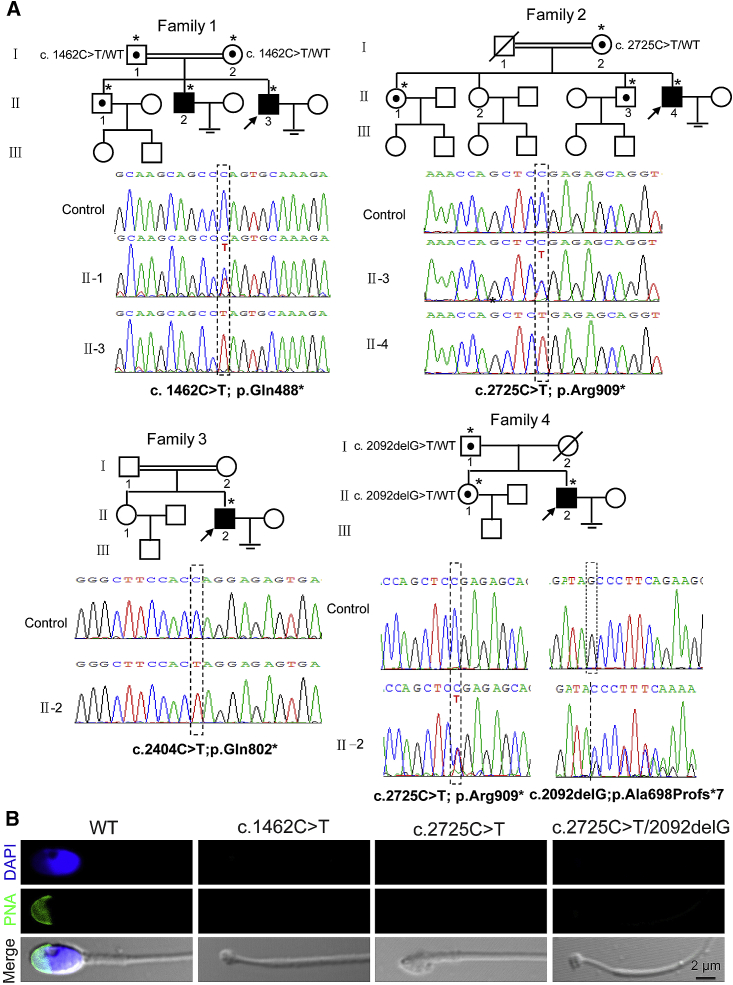
We identified a consanguineous family with two primary infertile brothers (Figure 1A, see Supplemental Note). The brothers had no previous record of significant illness and complete andrological examination revealed most of their semen parameters were normal but included acephalic spermatozoa (Figure 1B and Table S1). Whole-exome sequence analysis of one of the brothers implicated a homozygous nonsense mutation (Table S2), c.1462C>T (p.Gln488∗), in the coding region of PMFBP1 (GenBank: NM_031293.2), a gene specifically expressed in testis that encodes a sperm tail-associated protein (STAP), but its complete function had not yet been determined.25 To determine whether PMFBP1 is another acephalic spermatozoa syndrome-related gene, similar to SUN5 that we recently reported, we Sanger sequenced PMFBP1 in ten additional infertile men with acephalic spermatozoa syndrome and without SUN5 mutations.18 We identified two homozygous (c.2404C>T [p.Gln802∗]; c.2725C>T [p.Arg909∗]) mutations and one compound heterozygous (c.2725C>T [p.Arg909∗] and c.2092delG [p.Ala698Profs∗7]) mutation in three unrelated infertile men with acephalic spermatozoa syndrome (Figures 1A and 1B, Tables S1 and S2, see Supplemental Note). Segregation analysis could not be performed in the consanguineous family 3, with homozygous mutation c.2404C>T (p.Gln802∗), because parental DNA was not available. Other mutations identified in families 1, 2, and 4 were all heterozygous in corresponding individuals’ parents and segregated in these families as expected (Figure 1A). These mutations were absent in 100 unrelated normal men and in the data of the 1000 Genomes Project. These results imply that the PMFBP1 mutations might be responsible for acephalic spermatozoa syndrome in addition to SUN5 mutations.
Figure 1.
Pedigrees and PMFBP1 Mutations in Families Affected by Acephalic Spermatozoa Syndrome
(A) Pedigrees of the four families with inherited PMFBP1 mutations. Squares denote male family members, circles female family members, solid symbols affected members, open symbols unaffected family members, slashes deceased family members, equal signs infertility, and double lines a consanguineous marriage. The arrows indicate the index man. Individuals with an asterisk were Sanger sequenced.
(B) The spermatozoon are headless in the ejaculate of the PMFBP1 mutant men. Peanut agglutinin (PNA) staining (green) was performed in wild-type and PMFBP1 mutant spermatozoon. Nuclei were stained with DAPI (blue).
Pmfbp1-Deficient Mice Produce Acephalic Spermatozoa
To investigate whether PMFBP1 mutations are a main cause of acephalic spermatozoa syndrome, we generated Pmfbp1-knockout founder mice by applying the CRISPR-Cas9 system that targeted exon 3 and exon 10 of the Pmfbp1 gene (Figure S1). The PMFBP1 protein was completely absent in the Pmfbp1−/− testis compared with Pmfbp1+/− and wild-type (WT) testes (Figure 2A), indicating that homozygous mice were Pmfbp1 null. Pmfbp1−/− mice were viable and reached adulthood. The Pmfbp1−/− male mice failed to produce offspring (Figure 2B), while Pmfbp1-deficient female mice with normal follicle development were fertile (Figure S2), suggesting that Pmfbp1-deficient male mice are infertile.
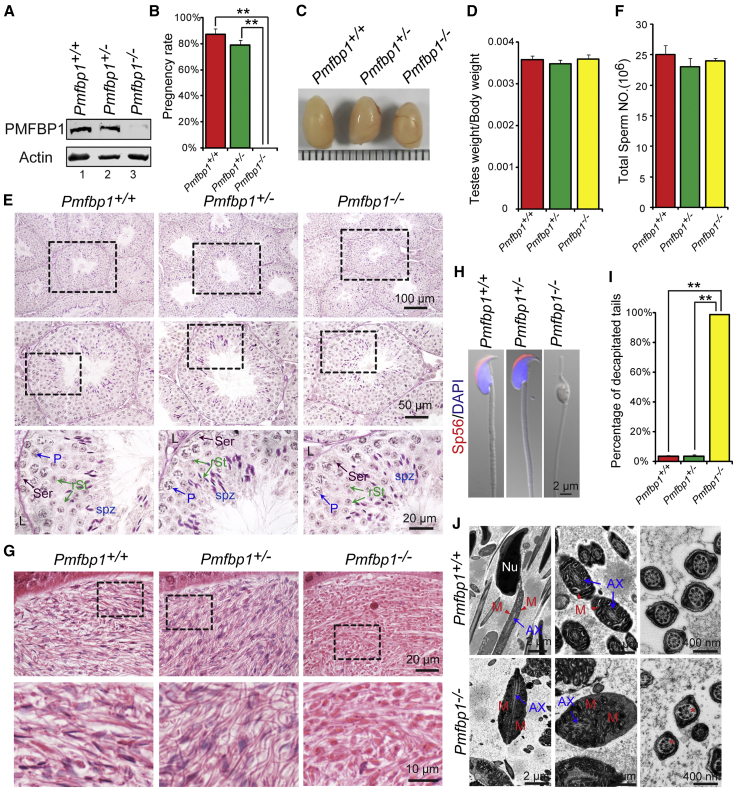
Figure 2.
The Disruption of Pmfbp1 in Mice Leads to Male Infertility due to the Production of Acephalic Spermatozoa
(A) The PMFBP1 protein was completely absent in the Pmfbp1−/− testis. Immunoblotting of PMFBP1 was performed in WT, Pmfbp1+/−, and Pmfbp1−/− testes. Actin served as a loading control.
(B) Pmfbp1−/− male mice were completely infertile. The fertility assessment experiments were performed in WT, Pmfbp1+/−, and Pmfbp1−/− male mice. The pregnancy rate: WT is 87.50% ± 4.17%, Pmfbp1+/− is 79.22% ± 3.22%, and Pmfbp1−/− is 0.00% ± 0.00%. Data are presented as mean ± SEM. ∗∗p < 0.01.
(C) The size of the testes was not altered in the Pmfbp1−/− and Pmfbp1+/− mice.
(D) Quantification ratio of testis weight/body weight in WT, Pmfbp1+/−, and Pmfbp1−/− male mice. WT 3.58 ± 0.10 × 10−3, Pmfbp1+/− 3.49 ± 0.08 × 10−3, and Pmfbp1−/− 3.60 ± 0.10 × 10−3. Data are presented as mean ± SEM.
(E) The histomorphology of Pmfbp1−/− seminiferous tubules were similar to the control groups as shown by PAS staining of testes in WT, Pmfbp1+/−, and Pmfbp1−/− mice. Abbreviations: L, Leydig cells; Ser, Sertoli cells; P, pachytene spermatocytes; rSt, round spermatid; spz, spermatozoa.
(F) The sperm counts in the caudal epididymis are similar in each group. WT 25.08 ± 1.46 × 106, Pmfbp1+/− 23.07 ± 1.36 × 106, and Pmfbp1−/− 24.03 ± 0.42 × 106. Data are presented as mean ± SEM.
(G) Fewer sperm heads are present in Pmfbp1−/− epididymis. The H&E staining of the caudal epididymis of WT, Pmfbp1+/−, and Pmfbp1−/− mice were shown. The spermatozoa within the Pmfbp1−/− caudal epididymis appear to be stained less with hematoxylin compared with those in the Pmfbp1+/− and WT epididymis.
(H) The Pmfbp1-null spermatozoa are headless. Single-sperm immunofluorescence analysis for the acrosome-specific marker sp56 (red) was performed using WT, Pmfbp1+/−, and Pmfbp1−/− spermatozoa. Nuclei were stained with DAPI (blue).
(I) Percentage of decapitated tails in WT, Pmfbp1+/−, and Pmfbp1−/− caudal epididymis. In WT and Pmfbp1+/− mice, 3.60% ± 0.62% and 3.73% ± 0.76% of spermatozoa were headless, whereas 98.74% ± 0.21% were headless in Pmfbp1−/− mice. Data are presented as mean ± SEM. ∗∗p < 0.01.
(J) Ultrastructure of WT and Pmfbp1−/− caudal epididymis showing that the Pmfbp1-null spermatozoa had no sperm head and contained a residual droplet of cytoplasm at the top of the flagellum with misarranged mitochondria inside. Abbreviations: Nu, nuclear; M, mitochondrion; AX, axoneme. The asterisk indicates the missing microtubule doublets of axoneme in Pmfbp1-null spermatozoa.
To explore the cause of the male infertility, we examined the Pmfbp1-deficient male mice at gross and histological levels. The testis size and weight of Pmfbp1−/− mice had no significant differences to those of Pmfbp1+/− and WT mice (Figures 2C and 2D). The histological examination by Periodic Acid-Schiff (PAS) staining revealed that the seminiferous tubules of Pmfbp1−/− mice were also similar to the control groups, and all of the components in the seminiferous epithelium could be detected in Pmfbp1−/− testis (Figure 2E). Since the intact acrosome is essential for male fertility,26, 27 we examined acrosome morphology at different steps of spermatids in Pmfbp1−/− mice by PAS staining and found that the acrosome morphology in Pmfbp1-deficient spermatids was normal (Figure S3A). The immunofluorescent staining of the acrosome-specific marker sp56 and TGN38 (the Golgi apparatus-derived vesicles marker) also revealed that the proacrosomal vesicles could be transported normally and fused to the proacrosomal structure (Figure S3B), indicating that the disruption of Pmfbp1 has no influence on the acrosome biogenesis.
Next, we counted the total sperm number in the caudal epididymis and found that no significant difference existed among the Pmfbp1−/−, Pmfbp1+/−, and WT mice (Figure 1F). However, the spermatozoa within the Pmfbp1−/− caudal epididymis appeared to be stained less with hematoxylin compared with those in the Pmfbp1+/− and WT epididymis, suggesting the absence of sperm heads in the Pmfbp1-deficient epididymis. Furthermore, single-sperm immunofluorescence revealed that Pmfbp1-null spermatozoa were negative for both sp56 and DAPI staining (Figures 1H and 1I), indicating the absence of both acrosome and nucleus. Quantitative analyses showed that the decapitated tails accounted for 98.74% ± 2.13% of the total sperm cells in the Pmfbp1−/− caudal epididymis (Figure 2I). Thus, the Pmfbp1 deficiency might lead to acephalic spermatozoa in mouse. To further confirm this phenomenon, we performed transmission electron microscopy (TEM) analysis of the Pmfbp1−/− and WT caudal epididymis. As shown in Figure 2J, the Pmfbp1-null spermatozoa had no sperm head and contained a residual droplet of cytoplasm at the top of the flagellum with misarranged mitochondria inside. In addition, the axoneme of decapitated tails in Pmfbp1-deficient mice was also abnormal (Figure 2J). Therefore, the Pmfbp1-null spermatozoa are actually acephalic spermatozoa, which may be responsible for the Pmfbp1−/− male mice infertility and consistent with PMFBP1 mutant individuals.
PMFBP1 Is Essential for the Attachment of the Coupling Apparatus to the Sperm Head
To investigate the pathogenic mechanisms of Pmfbp1 deficiency-caused acephalic spermatozoa syndrome, we first examined where the sperm head detaches from flagellum in Pmfbp1-deficient mice. Quantitative analyses showed that the ratios of decapitated tails in the Pmfbp1−/− corpus and caput of the epididymis were similar to those in the caudal epididymis (Figures 3A and 3B), indicating that the separation of the sperm head from the flagellum may occur within the seminiferous tubules. For further confirmation, we carefully examined the stages of spermiogenesis in Pmfbp1−/− and WT testes by PAS staining. We found that the mature sperm head could still be detected at stages IX–X in Pmfbp1-deficient testes, while mature spermatozoa were released at stage VIII in WT testes (Figure S4), indicating that the Pmfbp1-null sperm head and tail might break apart during spermiation. Furthermore, in the Pmfbp1-deficient testes, the orientation of the sperm head at stages V–VIII was oriented toward the lumen of the seminiferous tubules, but not the basement membrane, which might be caused by the sperm head detachment from flagellum during spermiogenesis (Figure S4). The detached sperm heads might be resolved within the seminiferous tubules, as some destroyed sperm heads were observed to be wrapped by the membrane at stages IX–X in Pmfbp1−/− testes (Figure S4). Thus, the connection between sperm head and tail might be destroyed during spermiogenesis in Pmfbp1-deficient testes.
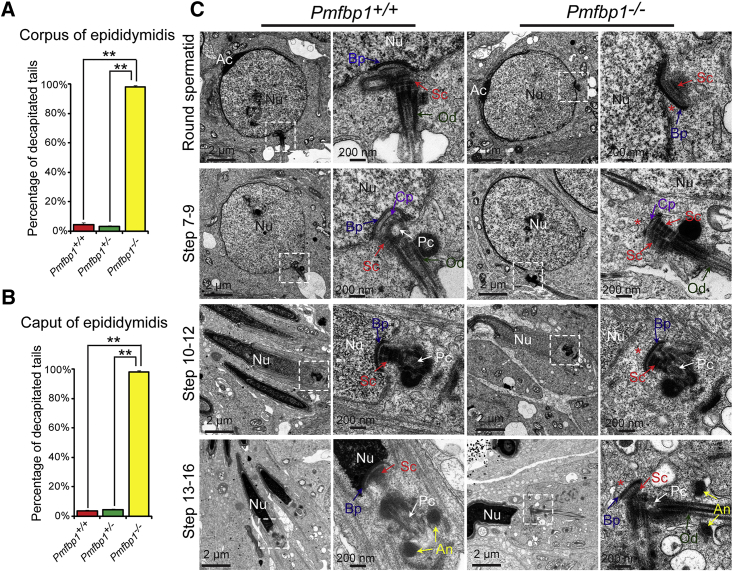
Figure 3.
PMFBP1 Is Essential for the Attachment of the Coupling Apparatus to the Caudal Portion of the Sperm Nucleus
(A and B) The percentage of decapitated tails in WT, Pmfbp1+/−, and Pmfbp1−/− corpus and caput epididymides. In WT, Pmfbp1+/−, and Pmfbp1−/− corpus epididymides, 4.69% ± 1.15%, 3.37% ± 0.59%, and 97.98% ± 1.06% of spermatozoa were headless (A). In WT, Pmfbp1+/−, and Pmfbp1−/− caput epididymides, 3.74% ± 0.77%, 4.64% ± 0.64%, and 98.15% ± 1.03% of spermatozoa were headless (B). Data are presented as mean ± SEM. ∗∗p < 0.01.
(C) The coupling apparatus could not be tightly attached to the sperm head in Pmfbp1-null mice. TEM analyses of the stepwise development of the coupling apparatus were performed in WT and Pmfbp1−/− testes. The coupling apparatus can be assembled in both WT and Pmfbp1−/− spermatid, but the coupling apparatus could not be tightly attached to the sperm head in Pmfbp1-null spermatids. The asterisk indicates the gap between the nuclear (Nu) envelope and the basal plate (Bp). Abbreviations: Nu, nuclear; Ac, acrosome; Bp, basal plate; Cp, capitulum; Sc, segmented column; Pc, proximal centriole; An, annulus; Od, outer dense fibers.
The flagellum is anchored to the sperm head through the head-tail coupling apparatus (HTCA), a centrosome-based structure consisting of two cylindrical microtubule-based centrioles and associated components.28, 29 To verify whether the destroyed sperm head to tail connection in Pmfbp1-deficient testes is caused by the defects in the development of the HTCA, we examined the ultrastructure of the spermatids in Pmfbp1−/− testes by TEM. In Pmfbp1-deficient round spermatids, the ultrastructure of the HTCA was fully assembled and consisted of a well-defined segmented column (Sc), capitulum (Cp), and basal plate (Bp) (Figure 3C), similar to the control group’s HTCAs. However, the coupling apparatus in Pmfbp1-deficient mice could only partially connect with the sperm nucleus in the implantation fossa (Figure 3C, top). Along with the elongation of the spermatid in Pmfbp1−/− testes, the unstable connection between the coupling apparatus and the sperm nucleus was destroyed, resulting in the separation of the coupling apparatus from the sperm nucleus (Figure 3C). Therefore, these results suggest that Pmfbp1 is essential for the tight attachment of the coupling apparatus to the caudal portion of the sperm head, and the disruption of Pmfbp1 leads to the coupling apparatus detachment from the sperm nucleus during spermiogenesis, causing acephalic spermatozoa syndrome in Pmfbp1-deficient mice.
PMFBP1 Is Localized at the Head-Tail Coupling Apparatus to Connect Sperm Head to Tail
PMFBP1 is specifically expressed in adult testis, located at the tail of elongated spermatids, and termed a STAP.25 To investigate the molecular mechanisms of PMFBP1 in the attachment of the coupling apparatus to the sperm nucleus, we characterized its precise localization during spermiogenesis using immunofluorescent staining in testis smears. We found that PMFBP1 was first expressed in the cytoplasm of round spermatids and later appeared in the implantation fossa region of the sperm nucleus during sperm head elongation and differentiation (Figure S5A). In step 12–16 spermatids and mature spermatozoa, PMFBP1 was predominantly located in the sperm head-to-tail connecting piece (Figures S5A and S5B). Further immunofluorescent staining of PMFBP1 and some other HTCA-related proteins, such as SUN5 and SPATA6 (spermatogenesis associated 6 [HGNC:18309, MIM: 613947]),17, 19 showed that PMFBP1 was located in the medium region between SUN5 and SPATA6 (Figure S5C and S5D). As SUN5 is a nuclear membrane protein connecting the coupling apparatus to the sperm nuclear envelope and SPATA6 is a structural protein specifically localized in the segmented columns and the capitulum of the coupling apparatus,17, 19 the sandwiched localization of PMFBP1 indicated that it might work as a scaffold protein to connect the coupling apparatus and sperm nuclear envelope. To verify this possibility, we determined the localization of SUN5 and SPATA6 in Pmfbp1-null spermatozoa and found that SUN5 was visible in the implantation fossa of the sperm nucleus (Figure 4A), whereas SPATA6 located in the residual droplet of cytoplasm at the top of the flagellum (Figure S6A). Thus, PMFBP1 is localized at the sperm head-to-tail connecting piece to join the coupling apparatus to the sperm nuclear envelope, supporting its function in sperm head and tail integrity. We next examined the interaction between SUN5, PMFBP1, and SPATA6 using glutathione S-transferase (GST)-pulldown and co-immunoprecipitation in cell culture (HEK293T cells). We found that the three proteins failed to interact with each other (Figure S7), indicating that some other HTCA components might be present to mediate the interaction between them.
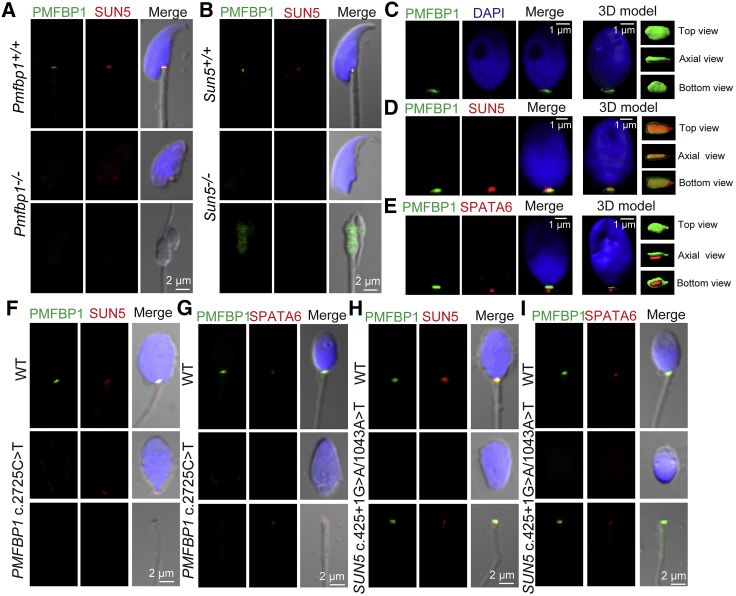
Figure 4.
PMFBP1 Cooperates with SUN5 and SPATA6 to Connect the Coupling Apparatus to the Sperm Head in Both Mouse and Human
(A) The disruption of Pmfbp1 had no obvious influence on the localization of SUN5. Single-sperm immunofluorescence analysis for PMFBP1 (green) and SUN5 (red) was performed in WT and Pmfbp1−/− spermatozoa. Nuclei were stained with DAPI (blue).
(B) Sun5 deficiency led to PMFBP1 detaching from sperm head and localizing in the residual droplet of cytoplasm at the top of the flagellum in the Sun5-null spermatozoa. Single-sperm immunofluorescence analysis for PMFBP1 (green) and SUN5 (red) was performed in WT and Sun5−/− spermatozoa. Nuclei were stained with DAPI (blue).
(C) PMFBP1 is localized in the sperm head-to-tail connecting piece in human. The single-sperm immunofluorescence analysis for PMFBP1 (green) was performed in human spermatozoa. Nuclei were stained with DAPI (blue). The three-dimensional model was reconstituted by IMARIS software.
(D and E) The sandwich-like structure of SUN5, PMFBP1, and SPATA6 in the coupling apparatus in human spermatozoa. Single-sperm immunofluorescence analysis of human spermatozoa for PMFBP1 (green) and SUN5 (red) is shown in (D). Immunofluorescence of PMFBP1 (green) and SPATA6 (red) analysis is shown in (E). Nuclei were stained with DAPI (blue). The three-dimensional models were reconstituted by IMARIS software.
(F and G) The mutation of PMFBP1 in human had no influence on the localization of SUN5, whereas the SPATA6 localized at the residual droplet of cytoplasm at the top of the flagellum. The single-sperm immunofluorescence analysis for PMFBP1 (green) and SUN5 (red) was performed in WT and PMFBP1 mutant spermatozoa (F). A similar immunofluorescence analysis of PMFBP1 (green) and SPATA6 (red) is shown in (G). Nuclei were stained with DAPI (blue).
(H and I) In SUN5 mutant spermatozoa, both PMFBP1 and SPATA6 were localized at the residual droplet of cytoplasm at the top of the flagellum. The single-sperm immunofluorescence analysis for the PMFBP1 (green) and SUN5 (red) was performed in WT and SUN5 mutant spermatozoa (H). A similar immunofluorescence analysis of PMFBP1 (green) and SPATA6 (red) is shown in (I). Nuclei were stained with DAPI (blue).
PMFBP1 Cooperates with SUN5 and SPATA6 to Maintain Sperm Head and Tail Integrity
After carefully comparing the ultrastructures of coupling apparatus between Pmfbp1−/− and Sun5−/− spermatozoa, we found that their phenotypes were consistent, with each showing the separation of the coupling apparatus from the sperm nuclear envelope (Figure 3C). These results indicate that PMFBP1 and SUN5 might work in a similar way to maintain sperm head and tail integrity. As the localization of SUN5 was not obviously influenced after Pmfbp1 deficiency, we speculated that PMFBP1 might work downstream of SUN5 to connect the coupling apparatus and the sperm nuclear envelope. To test this point, we determined the localization of PMFBP1 and SPATA6 by immunofluorescence in Sun5-null spermatozoa and found that both PMFBP1 and SPATA6 could be detected only in the residual droplet of cytoplasm at the top of the flagellum, but not in the implantation fossa (Figures 4B and S6C). Furthermore, the immunofluorescence of PMFBP1 in the Sun5-deficient testis smear also showed that PMFBP1 could not attach to the implantation fossa of the sperm nucleus (Figure S6B). Together, these results suggest that the disruption of Sun5 impairs the localization of PMFBP1 in the sperm head and that PMFBP1 may cooperate with SUN5 and SPATA6 to connect the coupling apparatus to the sperm head.
The Pathogenic Mechanisms of PMFBP1 in Acephalic Spermatozoa Syndrome Is Conserved in Human
Next, we examined whether the pathogenic mechanisms of Pmfbp1 deficiency in mouse model were conserved in PMFBP1 mutant individuals with acephalic spermatozoa syndrome. First, we determined the localization of PMFBP1 in normal human spermatozoa. Similar to the mouse spermatozoa, PMFBP1 was localized at the sperm head to tail connecting piece in human (Figure 4C). Immunofluorescence of PMFBP1, SUN5, and SPATA6 showed that all of the proteins were also visible around the HTCA of the human spermatozoa, and they presented as a sandwich-like structure with PMFBP1 located between SUN5 and SPATA6 (Figures 4D and 4E). These results indicate that the localization of PMFBP1, SUN5, and SPATA6 in the HTCA is conserved between mouse and human. Then, we determined the localization of SUN5 and SPATA6 in PMFBP1 mutant spermatozoa (c.2725C>T [p.Arg909∗]) and found that SUN5 was still localized in the implantation fossa region (Figure 4F), whereas SPATA6 was located at the top of the flagellum (Figure 4G), which are consistent with the phenomenon in mouse Pmfbp1-null spermatozoa. Thus, the human PMFBP1 mutation affects the attachment of the coupling apparatus to the sperm nuclear envelope. Further investigation revealed that the PMFBP1 and SPATA6 in the human SUN5-mutant spermatozoa (c.425+1G>A and c.1043A>T [p.Asn348Ile]) could be detected only at the top of the flagellum, but not in the implantation fossa (Figures 4H and 4I). Thus, the functional role of PMFBP1 in cooperating with SUN5 to connect the coupling apparatus to the sperm head is conserved in humans. Acephalic spermatozoa syndrome in PMFBP1 mutant men may also be caused by the detachment of the coupling apparatus from the sperm head during spermiogenesis.
Overcoming PMFBP1 Defect by ICSI
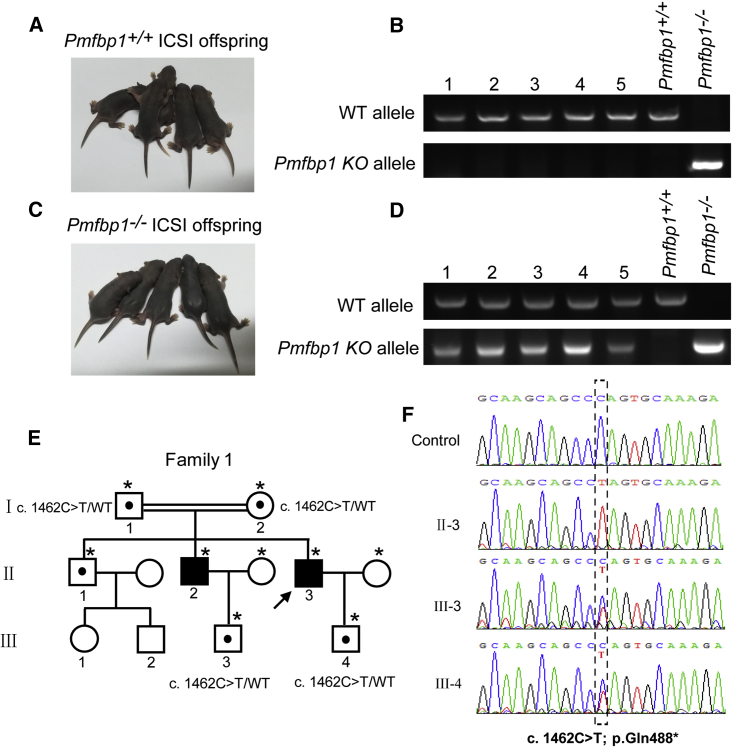
Our previous works revealed that SUN5 mutation-associated acephalic spermatozoa syndrome could be successfully overcome by intracytoplasmic sperm injection (ICSI).19 To examine whether the infertility caused by PMFBP1 mutant could also be overcome by using ICSI, we performed ICSI in Pmfbp1-deficient mice. Because very few sperm heads could be found in Pmfbp1−/− mouse caudal epididymis, we selected the tailless heads of Pmfbp1-null spermatozoa from the testes and injected them into WT oocytes, which resulted in healthy offspring of the Pmfbp1−/− mice (Figures 5A–5D). These results indicate that the infertility of Pmfbp1-deficient mouse could be overcome by ICSI. We then speculated that ICSI might also be suitable for patients with PMFBP1 mutation-associated acephalic spermatozoa syndrome. Although the proximal centriole is not required for mouse, it is essential for uniting sperm and oocyte pronuclei in the human being.30 The spermatozoa with abnormal head-tail junction from three men with PMFBP1 mutations were chosen for ICSI (Table S3). The partners of three men all became pregnant, and two healthy babies were born (Figures 5E and 5F and Table S3); the babies were further confirmed to be heterozygous for the PMFBP1 mutation (Figure 5F). These results suggest that PMFBP1 mutation-associated infertility could be successfully overcome by ICSI.
Figure 5.
Infertility Caused by PMFBP1 Mutations Could Be Overcome by ICSI
(A and B) Representative images (A) and genotypes (B) of the WT mouse ICSI offspring.
(C and D) Representative images (C) and genotypes (D) of the Pmfbp1−/− mouse ICSI offspring.
(E) Pedigree of family 1 with inherited PMFBP1 mutations, and the healthy babies (III-3 and III-4) of the infertility men after ICSI. Individuals with an asterisk were Sanger sequenced.
(F) Sequences of the PMFBP1 mutation sites of the representative individuals from each generation of family 1.
Discussion
Our previous work identified biallelic SUN5 mutations responsible for autosomal-recessive acephalic spermatozoa syndrome but only accounted for 47.06% of acephalic spermatozoa syndrome.18 These observations suggested the existence of unknown genes associated with acephalic spermatozoa syndrome. Recently, TSGA10 (testis specific 10 [HGNC:14927, MIM: 607166]) and BRDT (bromodomain testis associated [HGNC:1105, MIM: 602144]) have been identified to be associated with acephalic spermatozoa syndrome,8, 9 but these mutations have not been detected in other individuals or validated by animal models. In this study, we identified a homozygous nonsense mutation of PMFBP1 by employing WES in two infertile brothers with acephalic spermatozoa syndrome. Further Sanger sequencing of PMFBP1 in ten additional infertile men with acephalic spermatozoa syndrome and without SUN5 mutations revealed two homozygous mutations and one compound heterozygous mutation in three unrelated infertile men (Figures 1A and 1B). We produced Pmfbp1-deficient mouse models that strongly suggest the disruption of Pmfbp1 produces acephalic spermatozoa and impairs male fertility (Figure 2). Therefore, PMFBP1 is a novel acephalic spermatozoa syndrome-associated locus. In our cohort, we found that 72.00% (18/25) of acephalic spermatozoa syndrome is due to SUN5 and PMFBP1 mutations. By further examining recently reported acephalic spermatozoa syndrome-affected case subjects, we found that 70.00% (21/30) of acephalic spermatozoa men were caused by the mutation in either SUN5 or PMFBP1.8, 9, 18, 23, 31, 32 Therefore, SUN5 and PMFBP1 mutations are the main causes of acephalic spermatozoa syndrome. These two genes should be used as genetic markers for diagnosing acephalic spermatozoa syndrome in human.
PMFBP1 is specifically expressed in adult testis and locates at the sperm head to tail connecting piece between SUN5 and SPATA6 in both human and mouse (Figure 4). SUN5 is a nuclear membrane protein connecting the coupling apparatus to the sperm nuclear envelope, and SPATA6 is a structural protein of the coupling apparatus specifically localized in the segmented columns and the capitulum.17, 19 The sandwich-like structure of SUN5, PMFBP1, and SPATA6 reveals that PMFBP1 may work as a scaffold protein to connect the coupling apparatus and the sperm nuclear envelope. The ultrastructure of the coupling apparatus in Pmfbp1−/− testes showed that the basal plate of the coupling apparatus was intact (Figure 3C), suggesting that PMFBP1 is essential for the attachment of the basal plate to the sperm head. Nevertheless, PMFBP1 could not directly bind with SUN5 or SPATA6 (Figure S7), indicating that some other structural proteins are still needed to integrate SUN5, PMFBP1, and SPATA6 into the complete coupling apparatus. The homozygous nonsense mutation of PMFBP1 (c.2725C>T [p.Arg909∗]) leads to acephalic spermatozoa syndrome in human, which means that the C-terminal region of PMFBP1 is essential for the attachment of the coupling apparatus to the sperm head. However, the precise molecular mechanisms still need further investigation.
Previous studies have shown that acephalic spermatozoa have defects in the head-middle piece attachment, and ultrastructure analysis of acephalic spermatozoa in human and animals shows that the connecting piece of spermatozoa fails to form during late spermiogenesis.4, 5, 6, 18 According to the abnormal head-middle ultrastructure of acephalic spermatozoa in these reports, the case subjects were divided into two types: type I acephalic spermatozoa had decapitated heads that contained the proximal centriole that located in the implantation fossa with basal plate and type II acephalic spermatozoa had decapitated tails that contained the proximal and distal centriole.5 So far, most cases of acephalic spermatozoa belong to type II acephalic spermatozoa,3, 4, 5, 6, 7, 18 and our cases were no exception.18 In mouse, the ultrastructure analysis of Pmfbp1−/− and Sun5−/− testes also showed that the coupling apparatus completely detached from the sperm nuclear envelope (Figure 3C).19 Although the pathogenic mechanisms of PMFBP1 and SUN5 in acephalic spermatozoa syndrome are conserved between mouse and human, there were still some differences. The acephalic spermatozoa in Pmfbp1- and Sun5-deficient mice contained a residual droplet of cytoplasm at the top of the flagellum with distributed localization of SPATA6 or PMFBP1 (Figure S6A). In individuals with PMFBP1 or SUN5 mutations, however, these proteins could be detected at the top of the flagellum (Figures 4F–4I). These difference may be caused by the different ultrastructure of the HTCA in mouse and human, as the proximal centriole still presents in the HTCA of human mature spermatozoa,33 which may be helpful in maintaining the integrity of the coupling apparatus after detaching from the sperm nuclear envelope. Alternatively, the effect of human mutations on their functions are less severe than those of the mouse models, and the well-formed sperm heads detach from their tails after ejaculation or capacitation in those individuals, thus producing different kinds of acephalic spermatozoa.
Our current investigation revealed that men with PMFBP1 mutation-associated acephalic spermatozoa syndrome could achieve healthy offspring by ICSI, which is consistent with SUN5 mutation-associated individuals.19, 32 Thus, ICSI may be an effective therapeutic strategy to overcome infertility in acephalic spermatozoa syndrome. However, some important caveats should always be kept in mind during the treatment of individuals with acephalic spermatozoa syndrome. First, some acephalic spermatozoa look very much like globozoospermatozoa, and this type of pseudo-globozoospermatozoa needs to be carefully checked to be sure there is a nucleus in the head of the flagellum to avoid selecting the wrong type of sperm head. Second, as the centriole of human spermatozoa is essential for uniting sperm and oocyte pronuclei after fertilization,30 the spermatozoa with abnormal head-tail junction from an acephalic spermatozoa syndrome individual is suggested to be used for ICSI. If individuals with acephalic spermatozoa syndrome have no sperm heads in their ejaculate, the spermatozoa can be collected from the testes to perform ICSI. Third, once the acephalic spermatozoa syndrome individual has been diagnosed to contain PMFBP1 or SUN5 mutations, the mutations of these two genes should be screened in his partner before performing ICSI to avoid recessive homozygous mutations or compound heterozygous mutations in their offspring. Fourth, if his partner does unfortunately carry these mutations, female embryos are suggested to be selected during ICSI, because these mutations do not influence female in either mouse or human, while homozygous or biallelic mutations in the male may cause acephalic spermatozoa syndrome again.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank Tracey Baas for critical reading of the manuscript and Dr. Wei Yan for kindly gifting the SPATA6 antibody. This work was supported by the National key R&D program of China (grant no. 2016YFA0500901), the National Natural Science Foundation of China (81401251, 81572283, 31771501), and Scientific Research Foundation of the Institute for Translational Medicine of Anhui Province (SRFITMAP 2017zhyx29).
Published: July 19, 2018
Footnotes
Supplemental Data include seven figures, three tables, and Supplemental Note and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.06.010.
Contributor Information
Yunxia Cao, Email: caoyunxia6@126.com.
Wei Li, Email: leways@ioz.ac.cn.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
GenBank, https://www.ncbi.nlm.nih.gov/genbank/
HUGO Gene Nomenclature Committee, http://www.genenames.org/
OMIM, http://www.omim.org/
RefSeq, https://www.ncbi.nlm.nih.gov/RefSeq
WES protocol, https://www.agilent.com/cs/library/usermanuals/public/G7530-90005.pdf
Supplemental Data
References
- 1.Inhorn M.C., Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update. 2015;21:411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 2.Ray P.F., Toure A., Metzler-Guillemain C., Mitchell M.J., Arnoult C., Coutton C. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin. Genet. 2017;91:217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 3.Chemes H.E., Carizza C., Scarinci F., Brugo S., Neuspiller N., Schwarsztein L. Lack of a head in human spermatozoa from sterile patients: a syndrome associated with impaired fertilization. Fertil. Steril. 1987;47:310–316. doi: 10.1016/s0015-0282(16)50011-9. [DOI] [PubMed] [Google Scholar]
- 4.Perotti M.E., Giarola A., Gioria M. Ultrastructural study of the decapitated sperm defect in an infertile man. J. Reprod. Fertil. 1981;63:543–549. doi: 10.1530/jrf.0.0630543. [DOI] [PubMed] [Google Scholar]
- 5.Chemes H.E., Puigdomenech E.T., Carizza C., Olmedo S.B., Zanchetti F., Hermes R. Acephalic spermatozoa and abnormal development of the head-neck attachment: a human syndrome of genetic origin. Hum. Reprod. 1999;14:1811–1818. doi: 10.1093/humrep/14.7.1811. [DOI] [PubMed] [Google Scholar]
- 6.Baccetti B., Burrini A.G., Collodel G., Magnano A.R., Piomboni P., Renieri T., Sensini C. Morphogenesis of the decapitated and decaudated sperm defect in two brothers. Gamete Res. 1989;23:181–188. doi: 10.1002/mrd.1120230205. [DOI] [PubMed] [Google Scholar]
- 7.Porcu G., Mercier G., Boyer P., Achard V., Banet J., Vasserot M., Melone C., Saias-Magnan J., D’Ercole C., Chau C., Guichaoua M.R. Pregnancies after ICSI using sperm with abnormal head-tail junction from two brothers: case report. Hum. Reprod. 2003;18:562–567. doi: 10.1093/humrep/deg121. [DOI] [PubMed] [Google Scholar]
- 8.Li L., Sha Y., Wang X., Li P., Wang J., Kee K., Wang B. Whole-exome sequencing identified a homozygous BRDT mutation in a patient with acephalic spermatozoa. Oncotarget. 2017;8:19914–19922. doi: 10.18632/oncotarget.15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha Y.W., Sha Y.K., Ji Z.Y., Mei L.B., Ding L., Zhang Q., Qiu P.P., Lin S.B., Wang X., Li P. TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin. Genet. 2018;93:776–783. doi: 10.1111/cge.13140. [DOI] [PubMed] [Google Scholar]
- 10.Mendoza-Lujambio I., Burfeind P., Dixkens C., Meinhardt A., Hoyer-Fender S., Engel W., Neesen J. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 2002;11:1647–1658. doi: 10.1093/hmg/11.14.1647. [DOI] [PubMed] [Google Scholar]
- 11.Netzel-Arnett S., Bugge T.H., Hess R.A., Carnes K., Stringer B.W., Scarman A.L., Hooper J.D., Tonks I.D., Kay G.F., Antalis T.M. The glycosylphosphatidylinositol-anchored serine protease PRSS21 (testisin) imparts murine epididymal sperm cell maturation and fertilizing ability. Biol. Reprod. 2009;81:921–932. doi: 10.1095/biolreprod.109.076273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokuhiro K., Isotani A., Yokota S., Yano Y., Oshio S., Hirose M., Wada M., Fujita K., Ogawa Y., Okabe M. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 2009;5:e1000712. doi: 10.1371/journal.pgen.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liska F., Gosele C., Rivkin E., Tres L., Cardoso M.C., Domaing P., Krejcí E., Snajdr P., Lee-Kirsch M.A., de Rooij D.G. Rat hd mutation reveals an essential role of centrobin in spermatid head shaping and assembly of the head-tail coupling apparatus. Biol. Reprod. 2009;81:1196–1205. doi: 10.1095/biolreprod.109.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kierszenbaum A.L., Rivkin E., Tres L.L., Yoder B.K., Haycraft C.J., Bornens M., Rios R.M. GMAP210 and IFT88 are present in the spermatid golgi apparatus and participate in the development of the acrosome-acroplaxome complex, head-tail coupling apparatus and tail. Dev. Dyn. 2011;240:723–736. doi: 10.1002/dvdy.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K., Meinhardt A., Zhang B., Grzmil P., Adham I.M., Hoyer-Fender S. The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Mol. Cell. Biol. 2012;32:216–225. doi: 10.1128/MCB.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang K., Grzmil P., Meinhardt A., Hoyer-Fender S. Haplo-deficiency of ODF1/HSPB10 in mouse sperm causes relaxation of head-to-tail linkage. Reproduction. 2014;148:499–506. doi: 10.1530/REP-14-0370. [DOI] [PubMed] [Google Scholar]
- 17.Yuan S., Stratton C.J., Bao J., Zheng H., Bhetwal B.P., Yanagimachi R., Yan W. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc. Natl. Acad. Sci. USA. 2015;112:E430–E439. doi: 10.1073/pnas.1424648112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu F., Wang F., Yang X., Zhang J., Wu H., Zhang Z., Zhang Z., He X., Zhou P., Wei Z. Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am. J. Hum. Genet. 2016;99:942–949. doi: 10.1016/j.ajhg.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang Y., Zhu F., Wang L., Ouyang Y.C., Dong M.Z., Liu C., Zhao H., Cui X., Ma D., Zhang Z. Essential role for SUN5 in anchoring sperm head to the tail. eLife. 2017;6:6. doi: 10.7554/eLife.28199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H., Wan H., Li X., Liu W., Chen Q., Wang Y., Yang L., Tang H., Zhang X., Duan E. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:852–869. doi: 10.1038/cr.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess R.A., Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 2008;636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- 23.Shang Y., Yan J., Tang W., Liu C., Xiao S., Guo Y., Yuan L., Chen L., Jiang H., Guo X. Mechanistic insights into acephalic spermatozoa syndrome-associated mutations in the human SUN5 gene. J. Biol. Chem. 2018;293:2395–2407. doi: 10.1074/jbc.RA117.000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner D.K., Schoolcraft W.B. In vitro culture of human blastocysts. Towards Reproductive Certainty. 1999;9:378–388. [Google Scholar]
- 25.Ohuchi J., Arai T., Kon Y., Asano A., Yamauchi H., Watanabe T. Characterization of a novel gene, sperm-tail-associated protein (Stap), in mouse post-meiotic testicular germ cells. Mol. Reprod. Dev. 2001;59:350–358. doi: 10.1002/mrd.1041. [DOI] [PubMed] [Google Scholar]
- 26.De Braekeleer M., Nguyen M.H., Morel F., Perrin A. Genetic aspects of monomorphic teratozoospermia: a review. J. Assist. Reprod. Genet. 2015;32:615–623. doi: 10.1007/s10815-015-0433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrin A., Coat C., Nguyen M.H., Talagas M., Morel F., Amice J., De Braekeleer M. Molecular cytogenetic and genetic aspects of globozoospermia: a review. Andrologia. 2013;45:1–9. doi: 10.1111/j.1439-0272.2012.01308.x. [DOI] [PubMed] [Google Scholar]
- 28.Irons M.J. Synthesis and assembly of connecting-piece proteins as revealed by radioautography. J. Ultrastruct. Res. 1983;82:27–34. doi: 10.1016/s0022-5320(83)90094-1. [DOI] [PubMed] [Google Scholar]
- 29.Pasek R.C., Malarkey E., Berbari N.F., Sharma N., Kesterson R.A., Tres L.L., Kierszenbaum A.L., Yoder B.K. Coiled-coil domain containing 42 (Ccdc42) is necessary for proper sperm development and male fertility in the mouse. Dev. Biol. 2016;412:208–218. doi: 10.1016/j.ydbio.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schatten H., Sun Q.Y. The role of centrosomes in mammalian fertilization and its significance for ICSI. Mol. Hum. Reprod. 2009;15:531–538. doi: 10.1093/molehr/gap049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkhatib R.A., Paci M., Longepied G., Saias-Magnan J., Courbière B., Guichaoua M.R., Lévy N., Metzler-Guillemain C., Mitchell M.J. Homozygous deletion of SUN5 in three men with decapitated spermatozoa. Hum. Mol. Genet. 2017;26:3167–3171. doi: 10.1093/hmg/ddx200. [DOI] [PubMed] [Google Scholar]
- 32.Fang J., Zhang J., Zhu F., Yang X., Cui Y., Liu J. Patients with acephalic spermatozoa syndrome linked to SUN5 mutations have a favorable pregnancy outcome from ICSI. Hum. Reprod. 2018;33:372–377. doi: 10.1093/humrep/dex382. [DOI] [PubMed] [Google Scholar]
- 33.Manandhar G., Simerly C., Schatten G. Highly degenerated distal centrioles in rhesus and human spermatozoa. Hum. Reprod. 2000;15:256–263. doi: 10.1093/humrep/15.2.256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.