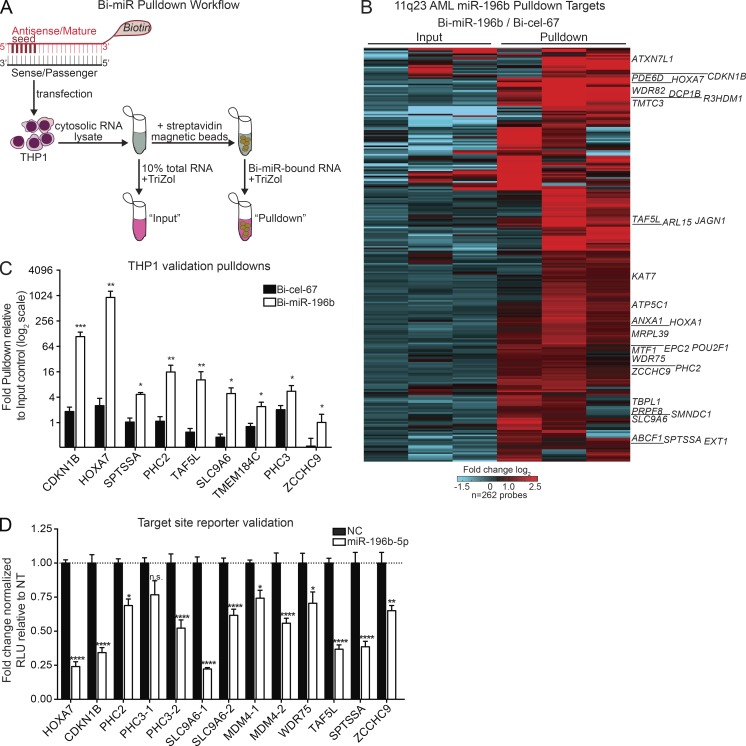
Figure 1.
Unbiased identification of direct miR-196b targets in human 11q23 AML. (A) Schematic of biotinylated miRNA-mimic (Bi-miR) pulldown approach. (B) Heat map of putative miR-196b target gene pulldowns enrichments from three independent pulldowns in 11q23 mutant THP1 AML cells. Displayed genes were enriched at least twofold in Bi-miR-196b pulldowns relative to matched Bi-cel-67 pulldowns in two of three experiments. The corresponding fold change Bi-miR-196b versus Bi-cel-67 inputs are shown for each gene. (C) Independent RT-qPCR validation of miR target mRNA pulldown. The average fold pulldown (± SEM) relative to matched input controls for each Bi-cel-67 control (black bars) and Bi-miR-196b mimic (white bars) in at least three independent experiments in THP1 cells. Statistical significance by paired t tests for each gene versus Bi-cel-67 control. (D) Functional miR target validation. Average fold change ± SEM repression by miR-196b (miR-196b-5p) relative to negative control (NC) of indicated miR-196b binding sites in three independent Luciferase reporter assay experiments. Multiple binding sites in the same gene are distinguished by “1” and “2”. Statistical significance was determined using a two-way ANOVA versus NC. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. n.s., not significant.