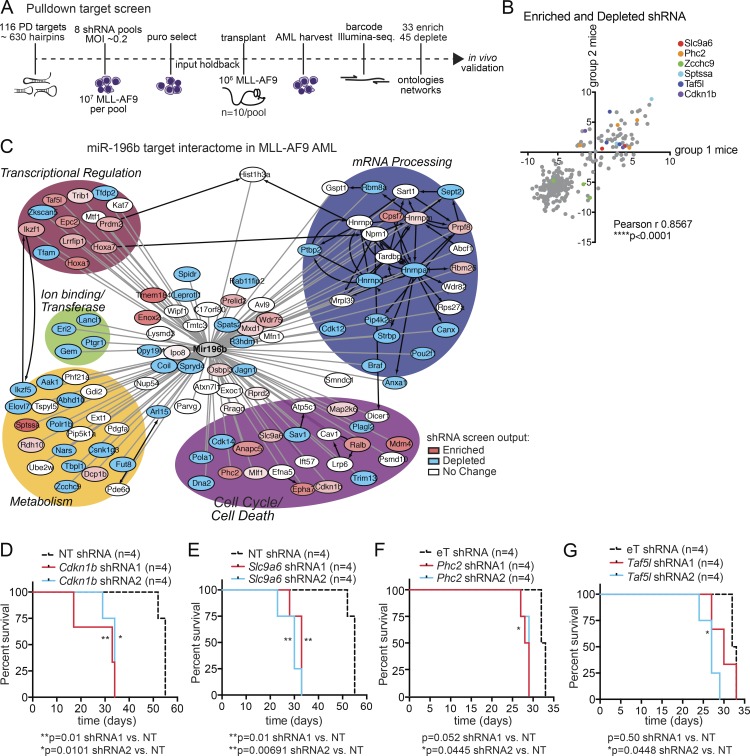
Figure 2.
An in vivo shRNA screen functionally dissects miR-196b networks in MLL-AF9 leukemogenesis. (A) Schematic overview. In vivo shRNA-positive selection screen of miR-196b-pulldown targets in primary MLL-AF9 leukemia. Leukemic splenocytes were transduced with eight different lentiviral shRNA pools against 116 miR-196b target genes and transplanted into recipient mice (n = 10 mice/pool). Sequencing of pretransplant pools (input) versus leukemic splenocytes (from moribund mice) identified 33 enriched and 45 deleted genes of at least two hairpins/gene over input control. (B) Replicate assay correlation. For each enriched and depleted hairpin, recipient mice were divided into two groups/pool (n = 4–5 mice/group/pool) and plotted. The relatedness between these two groups of mice was evaluated by Pearson correlation demonstrating that in vivo selection of hairpin activity is not stochastic (r = 0.8567). Examples of genes with enriched (Scl9a6, Phc2, Sptssa, Taf5l, and Cdkn1b) or depleted (Zcchc9) shRNA are color coded. (C) Network analysis of screen targets by gene ontology biological processes. RNAi screen enriched gene nodes (red), depleted gene nodes (blue), and unchanged gene nodes (white). Protein–protein interactions are denoted by black edges and putative miR-196b target transcript interactions experimentally identified in miR target pulldown assays are denoted by gray edges. (D–G) In vivo validation of positive selection of individual targets. Kaplan-Meier survival curves of mice transplanted with MLL-AF9 leukemia expressing individual shRNA hairpins against the indicated genes (n = 4 mice/group; 2 hairpins/gene). Non-targeting (NT) or EV (eT) were used as controls. miR-196b pulldown targets Cdkn1b (D), Slc9a6 (E), Phc2 (F), and Taf5l (G) accelerated leukemia lethality. Significant p-values are reported by Log-rank (Mantel-Cox) test for each hairpin compared with control. *, P ≤ 0.05; **, P ≤ 0.01; ****, P < 0.0001.