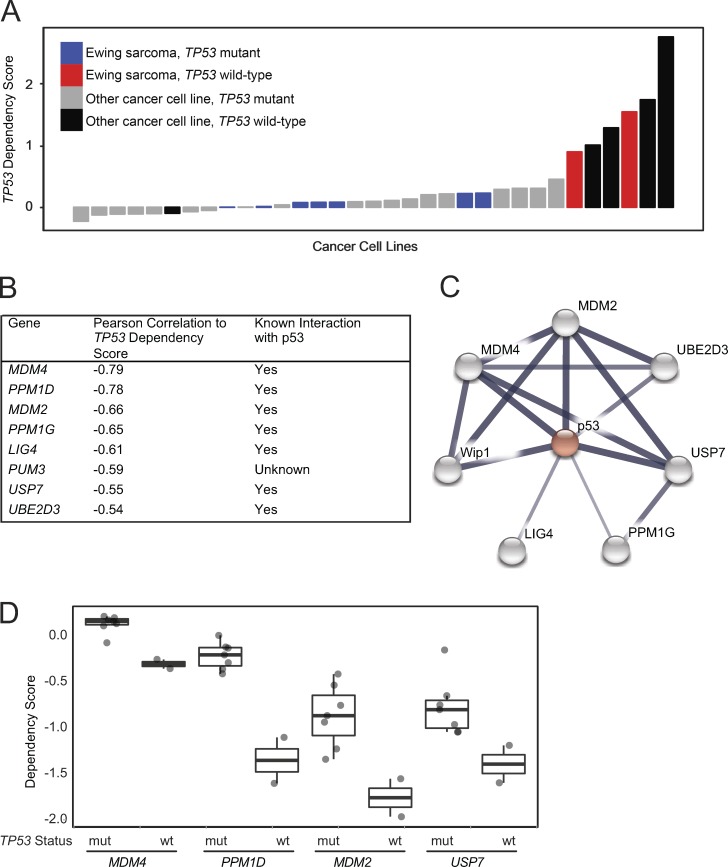
Figure 1.
Genome-scale CRISPR-Cas9 screen of 33 cancer cell lines identifies genetic vulnerabilities negatively correlated with TP53 dependency in TP53 wild-type lines. (A) Waterfall plot of TP53 dependency in 33 cancer cell lines shows positive dependency score in known TP53 wild-type cell lines consistent with the hypothesis that disruption of TP53 in these lines would lead to a proliferation advantage. Based on these data, 6 of 33 lines likely have a functional p53 pathway. A single cell line for which there is no documented TP53 mutation, PANC08.13, behaves like a TP53 mutant line, suggesting it has a nonfunctional p53 pathway. (B) Top eight anti-correlated genetic dependencies to TP53 dependency. (C) Seven of the top eight anti-correlated genes are connected to TP53 in the STRING database indicating putative protein–protein interactions. The widths of the edges correspond to the level of confidence in interactions (medium confidence STRING score of 0.4; high confidence STRING score of 0.7; highest confidence STRING score of 0.9). (D) MDM4, PPM1D, MDM2, and USP7 dependency scores in Ewing sarcoma cell lines in the CRISPR-Cas9 screen stratified by TP53 mutational status (mut, mutant; wt, wild-type).