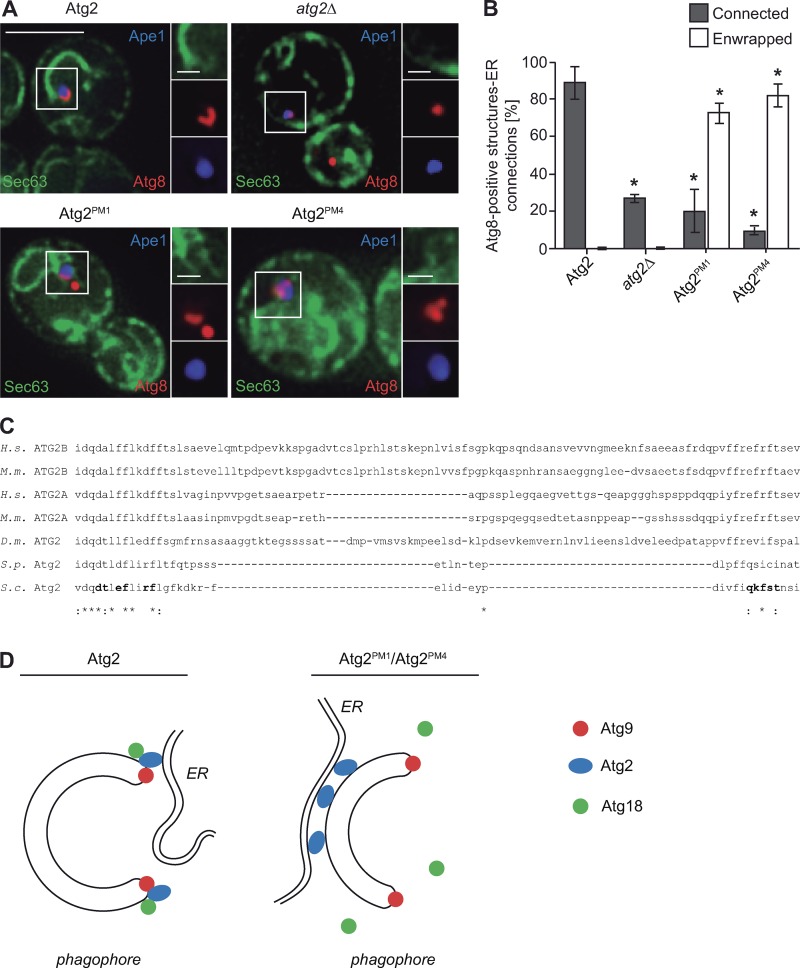
Figure 10.
Atg2 determines the contact sites between the phagophore and the ER. (A) Analysis of the ER–phagophore connection in cells generating giant Ape1 by fluorescence microscopy. The atg2Δ mutant expressing Sec63-GFP and mCherry-Atg8 (CUY10935) was transformed with both pDP105 and the pRS416 empty vector or a plasmid expressing Atg2 (pYCG_YNL242w), Atg2PM1 (pYCG_YNL242w_PM1), or Atg2PM4 (pYCG_YNL242w_PM4). The resulting strains were grown in SMD to an early log phase before to induce the formation of giant Ape1 as described in Materials and methods and to image the cells. Bars: (main images) 5 µm; (insets) 1 µm. (B) Quantification of the type of ER association to the mCherry-Atg8–positive phagophore in the experiment shown in C. Enwrapped defines all those situations when the ER was tethered to almost the entire surface of the phagophore, and Connected is when there was at least one point of contact between the ER and the phagophore. The graph represents the mean of three experiments ± SD. Asterisks indicate significant differences with cells expressing WT Atg2. (C) Conservation among species of the Atg2 residues involved in Atg9 binding. The amino acid sequence of S. cerevisiae (S.c.) Atg2 between residues 1,232 and 1,271 was aligned with that of Homo sapiens (H.s.) ATG2A and ATG2B, Mus musculus (M.m.) ATG2A and ATG2B, Drosophila melanogaster (D.m.), and Schizosaccharomyces pombe (S.p.) Atg2 using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/). The amino acids mutated in Atg2PM1 and Atg2PM4 are in bold. Asterisks indicate conservation of the residue, and colons designate similarity. (D) Left: Atg9 is confined at the extremities of the phagophore, where Atg2 also gets specifically concentrated by binding to this transmembrane protein. Atg9–Atg2 association also promotes the Atg18 recruitment, and collectively, these three factors play a key role in generating phagophore–ER contact sites at this location, although those appear to be preferentially generated at one of the two edges of the phagophore. Right: Inability of Atg2PM1 and Atg2PM4 to bind Atg9 impairs their targeting at the ends of the growing phagophore and Atg18 recruitment to this precursor structure. Redistribution of Atg2PM1 and Atg2PM4 on the phagophore surface leads to the formation of more extensive, wrongly positioned, and likely nonfunctional contact sites with the ER.