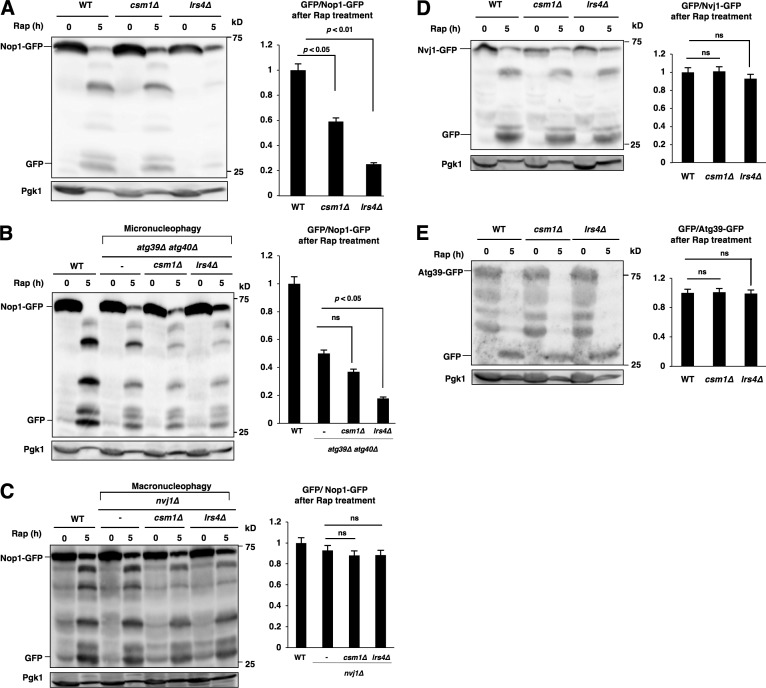
Figure 5.
Cohibin is required for nucleophagic degradation of nucleolar proteins. (A) Cells of strains BY4741 (WT), SCU2761 (csm1Δ), and SCU4885 (lrs4Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. Free GFP processed from GFP-tagged protein after rapamycin treatment was measured using ImageJ and quantified by calculating the ratio of cleaved free GFP to uncleaved full-length protein. (B) Cells of strains US356 (WT), SCU4575 (atg39Δ atg40Δ), SCU5037 (atg39Δ atg40Δ csm1Δ), and SCU5039 (atg39Δ atg40Δ lrs4Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (C) Cells of strains US356 (WT), SCU4514 (nvj1Δ), SCU5041 (nvj1Δ csm1Δ), and SCU5043 (nvj1Δ lrs4Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (D) Cells of strains SCU3287 (NVJ1-GFP), SCU4790 (csm1Δ NVJ1-GFP), and SCU4800 (lrs4Δ NVJ1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (E) Cells of strains SCU5143 (ATG39-GFP), SCU5145 (csm1Δ ATG39-GFP), and SCU5147 (lrs4Δ ATG39-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. The mean (± SD) was determined from three independent experiments, and relative values normalized against the value in control cells are shown. The p-values were calculated using two-tailed Student's t test.