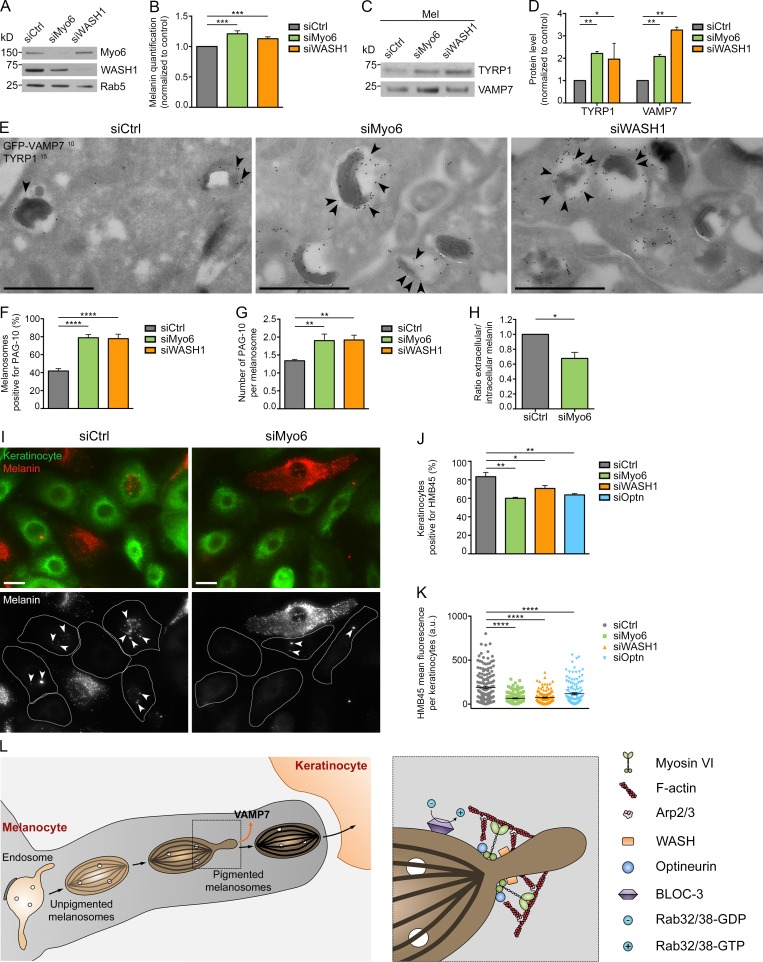
Figure 6.
Melanin production, cargo export, melanosome secretion, and transfer rely on Myo6, WASH, or optineurin. (A) Western blot of total lysates of control-, Myo6-, or WASH1-depleted MNT-1 cells probed with respective antibodies. (B) Intracellular melanin estimation of control-, Myo6-, or WASH1-depleted MNT-1 cells (n = 8 independent experiments; normalized to control). (C) Western blot of melanosome-enriched (Mel) fractions from control-, Myo6-, or WASH1-depleted MNT-1 cells probed with respective antibodies. (D) Quantification of TYRP1 and VAMP7 protein expression levels on Mel fractions in C and normalized to control (n = 2 independent experiments). (E) GFP-VAMP7–expressing MNT-1 cells treated with siCtrl, siMyo6, or siWASH1 were processed for ultrathin cryosectioning and double immunogold labeled for GFP (PAG 10 nm, arrowheads) and TYRP1 (PAG 15 nm). (F and G) Percentage of GFP-VAMP7+ melanosome per n cell on section (F) and relative number of gold particles per GFP-VAMP7+ melanosome (G; siCtrl, n = 7; siMyo6, n = 6; siWASH1, n = 7). (H) Ratio of extracellular to intracellular melanin content of control-, Myo6-, or WASH1-depleted MNT-1 cells treated with 30 µM forskolin (n = 3 independent experiments; normalized to control). (I) NHMs treated with control or Myo6 siRNAs were co-cultured with NHKs. Staining for HMB45 (red) or EGFR (green) identify melanin and NHM or NHK respectively. HMB45 staining within NHK (arrowheads) correspond to secreted and transferred melanin. (J and K) Quantification of the percentage of NHKs positive for at least one HMB45+ spot (J; n = 3 independent experiments) and of the mean HMB45 intensity per stained (n) NHK (K) when co-cultured with siCtrl, siMyo6, siWASH1 or siOptn NHM (siCtrl, n = 167; siMyo6, n = 147; siWASH1, n = 148; siOptn, n = 123). (L) Working model illustrating the roles of Myo6, optineurin, F-actin, WASH, Arp2/3, and BLOC-3 complexes in the recycling pathway from melanosomes. Molecular mass is in kilodaltons. Data are presented as the mean ± SEM. Bars: (E) 1 µm; (I) 10 µm. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (unpaired t test in B, D, F, G, and H; multiple comparisons ordinary one-way ANOVA in J and K).