Abstract
BACKGROUND AND OBJECTIVES
Genetic association studies have demonstrated that over 100 variants in target genes (including ADAM33) are associated with airway remodeling and hyper-responsiveness in different ethnic groups; however, this has never been evaluated in Arabic populations. The objective of this study was to determine whether ADAM33 polymorphisms that are associated with asthma in a population of asthmatic children from Saudi Arabia.
DESIGN AND SETTING
A cross-sectional pilot study comparing the polymorphisms of normal subjects and asthmatic patients from Saudi Arabia over a period of 1 year.
PATIENTS AND METHODS
One hundred and seven Saudi asthmatic children and 87 healthy Saudi children of 3–12 years old were assessed for allelic association of ADAM33 T1 (rs2280091), T2 (rs2280090), ST+4 (rs44707) and S1 (rs3918396) SNPs to asthma. Genotyping was done by real-time PCR, multiplex ARMS and PCR-RFLP.
RESULTS
T1 and T2 SNP genotype frequencies in asthmatic children were significantly different compared to controls (P<.05), indicating allelic association with asthma. The T1 A/G and G/G and the T2 A/G and A/A genotypes (P=.0013 and P=.008, respectively) but not S1 and ST+4, increased the risk of asthma when using the best fit dominant model. Strong linkage disequilibrium between T1 (rs2280091) and T2 (rs2280090) was observed (r2=0.83; D′=0.95; P<.001). The haplotype G-A-A-C was significantly more frequent in asthmatics, thus supporting the association of T1 G-allele and T2 A-allele with increased predisposition to asthma (P=.007).
CONCLUSIONS
T1 A/G and T2 G/A ADAM33 polymorphisms, but not S1 or ST+4, were significantly associated with asthma development in Saudi children, like those reported for white and Hispanic populations in the United States.
Asthma is a pulmonary disease characterized by intermittent narrowing of the small airways of the lungs, with subsequent reversible airflow obstruction, airway inflammation and bronchial hyper-responsiveness.1,2 Extensive epidemiological studies on asthma prevalence in different populations have shown a rise in asthma incidence since the 1960s and it is considered one of the most prevalent diseases in the world.3,4 In Saudi Arabia, 11% of the population suffers from asthma, while 4% to 23% of school students are asthmatic.5,6 Epidemiological studies indicate that many factors including age, gender, multiple genes and environmental factors influence the development of asthma. Several allelic association studies in different ethnic groups showed that more than 100 variants in target genes are linked with asthma, including ADAM33.7–9 The gene coding for ADAM33 (a disintegrin and metalloproteinase 33) is strongly associated with asthma susceptibility and bronchial hyper-responsiveness. 10,11 The ADAM33 gene is highly polymorphic, containing more than 70 single nucleotide polymorphisms (SNPs).11–13 At least 28 haplotype SNP pairs were shown to be significantly associated with asthma in United Kingdom and United States populations.11 Positive associations have also been reported between some of these polymorphisms and asthma in African-Americans and Hispanics.14 Although not without controversy, several SNPs in this gene have been shown to be associated with asthma susceptibility in other populations.15,16 In a Japanese cohort, the S2, T1, and T2 SNPs were significantly associated with asthma susceptibility.17 The ST+4 SNP was also linked to moderate and severe asthma in populations from the United Kingdom, the United States and Thailand.11,18 Furthermore, T1 and T2 were significantly associated with high total IgE and with asthma, respectively, in a population from Colombia.19 However, there is also lack of consistency in the results from several genetic association studies when tested in different populations. 20,21 In fact, the absence of association between ADAM33 polymorphisms including T1, T2 and S1, with asthma has been reported for various ethnic populations from Germany and the United States.10,22 Since no previous gene association studies have yet been reported on asthmatics from Saudi Arabia, it remained to be seen whether the ADAM33 polymorphisms could be associated with asthma in this population. The objective of this study was to examine in children from Saudi Arabia, the possible association of T1, T2, S1, and ST+4 SNPs with asthma susceptibility.
PATIENTS AND METHODS
One hundred and seven Saudi asthmatic children and 87 healthy Saudi children 3–12 years old were recruited at King Khalid University Hospital (KKUH) in Riyadh, Saudi Arabia. This study was approved by the College of Medicine Research Council (CMRC) Ethical Committee of the King Khalid University Hospital (KKUH). Informed written consent was obtained from the patients’ parents. Asthma diagnosis and severity assessment were determined based on symptoms, physician diagnosis (pediatric allergists or pulmonologists) following the criteria described by the American Thoracic Society. Demographic and clinical data for the asthmatics are presented in Table 1. The male to female ratio was approximately 2:1. The mean (SD) age for male and female children was similar (male, 7.4 [3.9] years old; female, 6.9 [3.0] years old). The mean age of onset in the male and female were also statistically similar (male, 53.8 [53.1] months; female, 42.3 [43.8] months) (P>.05). No significant differences were observed in the mean frequency of asthmatic exacerbations between the male and female children (P>.05). Among the clinical symptoms, cough, wheezing and shortness of breath were the major symptoms in both genders. Except for wheezing, which occurred more frequently in boys, all the other symptoms occurred at the same frequency in both genders. Approximately 60% of the patients (both male and female) were atopic. The severity of the disease differed significantly between both genders: the mild persistent disease was more frequent in females (84%) than in males (57%). Moderate-to-severe asthmatics were more frequent in males (41%) compared to females (13%). Over 90% of the patients were on medications, which included β2-adrenergic agonists, inhaled corticosteroids and anti-cholinergic agents. Leukotriene modifiers were used by more female patients (83.8%) than by male patients (67.1%).
Table 1.
Demographics, clinical data, medication and severity of the disease in asthmatic children patients.
| Asthmatic patients | Males (%) | Females (%) |
|---|---|---|
|
| ||
| Number of patients by gender | 70 (65.4) | 37 (34.6) |
| Mean of age (years) | 7.42 | 6.89 |
| Mean age of onset (months) | 53.8 | 42.3 |
| Mean frequency of asthma attack (months) | 3.9 | 3.6 |
| Cough | 68 (97.1) | 37 (100) |
| Wheezing a | 69 (98.6) | 31 (83.8) |
| Chest tightness | 63 (90) | 30 (81.1) |
| Breathlessness | 3 (4.3) | 5 (13.5) |
| Allergy | 44 (62.8) | 22 (59.5) |
| Drugs | ||
| β2-Adrenergic Agonists | 65 (92.8) | 35 (94.6) |
| Bronchodilators | 63 (90) | 35 (94.6) |
| Inhaled Corticosteroids | 65 (92.8) | 35 (94.6) |
| Anti-cholinergic agents | 65 (92.8) | 35 (94.6) |
| Leukotriene Modifiers | 47 (67.1) | 31 (83.8) |
| Disease Severityb | ||
| Mild Intermittent | 1 (1.4) | 1 (2.7) |
| Mild Persistent | 40 (57.1) | 31 (83.8) |
| Moderate Persistent | 22 (31.4) | 4 (10.8) |
| Severe Persistent | 7 (10) | 1 (2.7) |
P=.01,
P=.031 for the differences in the means between males and females.
For each parameter, quantities represent the number of subjects and its corresponding percentage value within parenthesis.
Four ADAM3 SNPs (T1, rs2280091; T2, rs2280090; S1, rs3918396; ST+4 (rs44707) were genotyped by real-time PCR using primers and TaqMan probes (Applied Biosystems, California, United States). The genotyping primers and TaqMan allele-specific probes are shown in Table 1. The probe for allele 1 was labeled with VIC dye and the probe for allele 2 was labeled with FAM dye. In this allelic discrimination assay, the presence of the two probes in each sample reaction (duplex) allows genotyping of two possible variants at the single base-pair nucleotide polymorphism and to discriminate for homozygotes (samples having only allele 1 or allele 2) and heterozygotes (samples having both allele 1 and allele 2). The T2 (rs2280090) SNP genotyping was confirmed by PCR-RFLP. Briefly, after PCR under standard reaction conditions, the amplified fragments were cut with NcoI (New England Biolabs, Beverly, Massachusetts, United States) and the digestion products were visualized on 4% agarose gels. The ST+4 (rs44707) SNP genotyping was performed by the multiplex tetra-primer amplification refractory mutation system PCR (ARMS-PCR), as described elsewhere.23 Briefly, both the wild type and the mutant alleles were amplified simultaneously by one pair of common outer primers flanking the SNP. The addition of two allele specific inner primers designed in opposite orientation and overlapping at the single base-pair nucleotide polymorphism allows the specific amplification of each allele. Since both products have different lengths (177bp for allele C; 138bp for allele A), they were clearly distinguished by agarose gel electrophoresis.
Allele and genotype frequencies for each SNP were obtained by allele and genotype counting. Chi-square tests were performed (P values <.05 were considered statistically significant) using SPSS software (version 17) (IBM Corp, Armonk, New York, United States). Hardy-Weinberg equilibrium tests, pairwise linkage disequilibrium and haplotype analyses were performed using SNPStats software (Institut Català d’Oncologia, Spain).24 To determine whether significant association between asthma and SNPs existed (genotypes and haplotypes), both crude analysis (nonadjusted) and adjusted for covariate gender were performed. Best fit genetic model tests (co-dominant, dominant, recessive and over-dominant) were performed using the chi-square and the odds ratios (OR) and 95% confidence intervals (CI) were calculated with the SNPStats software.24
RESULTS
T1 and T2 ADAM33 polymorphisms and asthma susceptibility in Saudi children
Fisher exact tests indicated that the T1, T2, ST+4 and S1 SNPs were distributed in Hardy-Weinberg proportions (P>0.05). A crude analysis (nonadjusted) indicated that only T1 and T2 SNPs were significantly associated with asthma. Since matching was not completely achieved on recruitment, we cannot rule out the possibility that such association analysis could be biased. Therefore, to eliminate possible residual con-founding by covariate gender, we present in Table 2 the association analysis with adjusted P values and odds ratios with 95% confidence intervals for genotypes. The association of T1 and T2 with asthma remained significant when the analysis was adjusted for covariate gender (Table 2). In the case of T1 SNP, the dominant model was the best fit effect model, based on the lowest Akaike (AIC=241) and Bayesian (BIC=251) index criteria and P value (P=.001), indicating that the presence of a single copy of G (with both the heterozygous A/G and the homozygous G/G having the same effect) is enough to increase the risk or predisposition to asthma (Table 2). Significant differences in the frequency of T1 (rs2280091) were observed in asthmatics when compared to healthy control groups (Table 3). The frequencies of the heterozygous A/G and the homozygous G/G T1 genotypes were significantly higher in the asthmatic group (49.5% and 11.3%, respectively), compared to the healthy group (32.9% and 5.9%, respectively) (P=.014). The homozygous A/A genotype was significantly more frequent in the healthy group (61.2 %) than in the asthmatic children (39.2%) (Table 3). The frequency of T2 (rs2280090) G/A genotype was significantly higher (48.9%) in asthmatic children than in healthy children (29.8%), whereas the ‘wild-type’ G/G genotype occurred at lower frequency in the asthmatics (43.6%), compared to healthy children (61.9%) (P=.031) (Table 3). The frequency of G/A and A/A combined were significantly higher in the asthmatics (56.4%), compared to the healthy group (38.1%) (Table 2). The dominant model was the best-fit genetic model of T2 SNP association to asthma, in which one copy of A (G/A and A/A having same effect) was enough to increase the risk or predisposition (P=.008) (Table 2). Concerning polymorphisms ST+4 (rs44707) and S1 (rs3918396t), no significant association with asthma was observed (P>.05) (Tables 2, 3).
Table 2.
Genotype frequency distribution of ADAM33 T1, T2, ST+4 and S1 polymorphisms and its association with asthma.
| SNP | Best Genetic Model | Genotype frequency, number (%) | OR (95% CI) | P | AIC (BIC) | ||
|---|---|---|---|---|---|---|---|
| Genotype | Healthy | Asthmatic | |||||
|
| |||||||
| T1 | Dominant | A/A | 53 (61.6) | 38 (39.6) | 1.00 | .001 | 241 (251) |
| rs2280091 | A/G-G/G | 33 (38.4) | 58 (60.4) | 2.70 (1.46–5.01)a | |||
| T2 | Dominant | G/G | 52 (61.9) | 41 (43.6) | 1.00 | .008 | 238 (247) |
| rs2280090 | G/A-A/A | 32 (38.1) | 53 (56.4) | 2.28 (1.22–4.23)a | |||
| ST+4 | NA | A/A | 32 (53.3) | 48 (48.5) | 1.00 | 0.49 | 211 (221) |
| rs44707 | A/C-C/C | 28 (46.7) | 51 (51.5) | 1.26 (0.66–2.43) | |||
| S1 | NA | C/C | 80 (97.6) | 90 (93.8) | 1.00 | 0.28 | 244 (254) |
| rs3918396 | C/T | 2 (2.4) | 6 (6.2) | 2.38 (0.46–12.37) | |||
Analysis of allele frequencies adjusted by covariate gender, showing odds ratio (OR) and 95% confidence interval (CI) for the best fit model of SNP association with asthma. For each SNP, 4 models were tested, using the most frequent homozygous genotype as reference. The OR and CI for only the best fit genetic model are shown, selected by their lowest index criteria values (AIC and BIC). NA means None Available.
Table 3.
Genotype frequencies (%) of T2, T1, ST+4 and S1 polymorphisms in ADAM33 gene.
| SNP | Healthy (H) or Asthmatic (A) | Genotype frequency (%) | P | ||
|---|---|---|---|---|---|
|
| |||||
| T1 rs2280091 |
A/A | A/G | G/G | ||
| H | 61.2 | 32.9 | 5.9 | .014 | |
| A | 39.2 | 49.5 | 11.3 | ||
| T2 rs2280090 |
G/G | G/A | A/A | ||
| H | 61.9 | 29.8 | 8.3 | .031 | |
| A | 43.6 | 48.9 | 7.4 | ||
| ST+4 rs44707 |
A/A | A/C | C/C | ||
| H | 53.3 | 46.7 | 0 | .53 | |
| A | 49.0 | 45.9 | 5.1 | ||
| S1 rs3918396 |
C/C | C/T | T/T | ||
| H | 97.6 | 2.4 | 0 | .22 | |
| A | 93.8 | 6.3 | 0 | ||
P value represents the differences in the genotype frequencies between healthy subjects (H) and asthmatic patients (A).
Interaction of T1 and T2 SNPs genotype frequencies with gender and susceptibility to asthma
We analyzed whether single SNP-to-gender interactions could alter the risk of asthma (Table 4). In the tested model where genotypes are nested within genders, we observed ORs with statistical significance in females for T1 G/G genotype (OR=13.6; 95% CI=1.5–127.6) and in males for T1 A/G (OR=3.6; 95% CI=1.4–8.9) (global interaction P=.054). These results highlight significant differences in genotype frequencies of T1 G/G (18.2% for asthmatic females vs. 2.3% for healthy females) and of T1 A/G (49.2% in asthmatic males vs. 22.0% for healthy males) (Table 4). Concerning T2 SNP, the G/A genotype reached statistical significance (OR=2.9; 95% CI=1.2–7.2), thus indicating an association to increased risk of asthma in boys, but not in girls (Table 4). Chi-square tests for T2 SNP confirmed the above result: the frequency of heterozygous G/A genotype was significantly higher in asthmatic boys (47.6%) than in healthy boys (22.5%; P=.022). In the case of ST+4 and S1 SNPs, no significant interactions between genotypes and genders were observed.
Table 4.
Interaction analysis of single SNP-gender, with genotypes nested within genders.
| SNP | Gender | Genotype | Genotype Frequency, number & (%) | OR (95% CI) | Global Interaction P value |
|
|---|---|---|---|---|---|---|
| Healthy | Asthmatics | |||||
|
| ||||||
| T1 rs2280091 |
F | A/A | 25 (55.5) | 11 (33.3) | 1 | .054 |
| A/G | 19 (42.2) | 16 (48.5) | 1.91 (0.7–5.10 | |||
| G/G | 1 (2.3) | 6 (18.2) | 13.6 (1.5–127.6)a | |||
| M | A/A | 28 (68.3) | 27 (42.9) | 1 | ||
| A/G | 9 (22.0) | 31 (49.2) | 3.6 (1.4–8.9)a | |||
| G/G | 4 (9.8) | 5 (7.9) | 1.3 (0.3–5.4) | |||
| T2 rs2280090 |
F | G/G | 26 (59.1) | 11 (35.5) | 1 | .13 |
| G/A | 16 (36.4) | 16 (51.6) | 2.36 (0.9–6.3) | |||
| A/A | 2 (4.5) | 4 (12.9) | 4.7 (0.7–29.7) | |||
| M | G/G | 26 (65.0) | 30 (47.6) | 1 | ||
| G/A | 9 (22.5) | 30 (47.6) | 2.9 (1.2–7.2)a | |||
| A/A | 5 (12.5) | 3 (4.8) | 0.5 (0.1–2.4) | |||
| ST+4 rs44707 |
F | A/A | 14 (45.2) | 17 (50.0) | 1 | .41 |
| A/C | 17 (54.8) | 14 (41.2) | 0.7 (.2–1.8) | |||
| C/C | 0 (0.0) | 3 (8.8) | NA | |||
| M | A/A | 18 (62.1) | 31 (47.7) | 1 | ||
| A/C | 11 (37.9) | 32 (49.2) | 1.7 (0.7–4.2) | |||
| C/C | 0 (0.0) | 2 (3.1) | NA | |||
| S1 rs3918396 |
F | C/C | 42 (97.7) | 32 (97.0) | 1 | .61 |
| C/T | 1 (2.3) | 1 (3.0) | 1.3 (0.1–21.8) | |||
| T/T | 0 (0.0) | 0 (0.0) | NA | |||
| M | C/C | 38 (97.4) | 58 (92.1) | 1 | ||
| C/T | 1 (2.6) | 5 (7.9) | 3.3 (0.4–29.1) | |||
| T/T | 0 (0.0) | 0 (0.0) | NA | |||
indicates significance of odds ratios (OR) and confidence intervals (95% CI) for heterozygous and less frequent homozygous genotypes, compared to the most frequent homozygous genotype
Pairwise linkage disequilibrium and haplotype frequencies
The possibility that two polymorphisms could be associated with an increased risk of asthma was analyzed by linkage disequilibrium (LD) and haplotype assessment. Data analysis showed a strong LD between T1 (rs2280091) and T2 (rs2280090), as measured by high D′ (0.95), elevated r2 (0.83) and X2 (286.6) (P<.001) (Table 2). ST+4 and S1 SNPs were not significantly linked to any of the above markers (P>.05). On the basis of LD, we analyzed the haplotype frequencies and their association to asthma. Table 5 describes the most frequent haplotypes, which together represent a cumulative frequency of 0.97. The ORs and 95% CIs were obtained from the estimated haplotype frequencies, taking as a reference the most common haplotype, [H1 A-G-A-C]. As expected, the wild-type haplotype [H1 A-G-A-C] was more frequent in the healthy group (0.52) than in the asthmatic group (0.35) (Table 5). Importantly, the haplotype H2 [G-A-A-C] was significantly more frequent in the asthmatic group (0.31) than in the healthy group (0.21) (OR=2.4; 95% CI=1.2–4.6), suggesting an association with asthma (P=.009). Other frequent haplotypes (H3, H4 and H5; Table 5) showed no significant association with asthma (P>.05).
DISCUSSION
Asthma is one of the most prevalent chronic diseases in the world.3,4 More than 300 million individuals suffer from this disease, and the prevalence of asthma continues to be on the rise in several populations.3,4,25 Asthma occurs at a high prevalence (10%–23%) in Saudi Arabia. 5,6 Genetic studies of asthma in the Saudi population have only recently been initiated and limited information is available. To our knowledge, this is the first re port on the association of ADAM33 polymorphisms with asthma in a Saudi population. ADAM33 has a strong relevance to the pathogenesis of asthma, mainly in relation to airway remodeling and bronchial hyper-responsiveness. 10,11 Several studies have indicated wide variations in the frequency of association of different ADAM33 SNPs with asthma.9,22,26 Among 37 polymorphisms identified in ADAM33, six were found to be significantly associated with asthma in a combined study on the United Kingdom/United States populations, including T2, T1, and S1.11
In the present study, T1 and T2 SNPs were found to be linked with asthma susceptibility in the Saudi population, thus confirming the findings of previous reports in other ethnic groups.17,19,27 The T2 G/A and the T1 A/G polymorphisms occur both in the cytoplasmic domain of ADAM33. Experimental evidence suggests that these modifications potentially alter the protein’s intracellular signaling function and may result in increased fibroblast and smooth muscle proliferation, which are characteristic features of airway remodeling in asthmatics.10,11,13,14,28 The analyses showed that over the four genetic models tested, the dominant model explained best the genotype frequencies of T1 and T2 markers. In this model, a single copy of the mutant G allele (T1) is enough to increase the risk or predisposition to asthma; therefore, the A/G and G/G genotypes will have the same effect. The same applies to the T2 SNP, in which the dominant model indicated that the presence of the mutant A allele increases the risk of asthma. The lack of association for ST+4 and S1 polymorphisms could be explained at least in part, by the very low frequency of the ST+4 homozygous C/C genotype and by the complete absence of the S1 T/T genotype in Saudi children. One possibility is that these results could have been biased due to the relatively small sample populations in this work. These results should be replicated in future studies with a larger Saudi population. Nevertheless, the lack of association of S1 and ST+4 SNPs with asthma in our study goes in agreement with several reports stating that these mutations do not significantly increase the risk or susceptibility to asthma.10,22,28 However, other studies found an association of S1 SNP with reduced lung function in children, when the recessive model was considered.10,11,27,29
The gene-environment interaction influencing the risk of asthma is complex and still not completely understood. Gender is one important environmental factor influencing asthma susceptibility; in childhood asthma, there are about 2-fold more cases of boys affected than girls, and this situation reverses at a later age.30,31 When we analyzed the genotype-gender interaction with risk or susceptibility to asthma, significant differences were observed in the T1 and T2 SNPs interactions with covariate gender. The T1 A/G and the T2 G/A genotypes were associated with increased risk of asthma in boys. In contrast, only the low frequency T1 G/G genotype was significantly associated with this risk in girls. These results suggest that boys in this cohort may be genetically more susceptible than girls. These results are in agreement with some clinical data of this cohort: mild asthma was more frequent in girls (84%) than boys (57%), whereas moderate-severe asthma was more frequent in boys (41%) than girls (13%); wheezing was also more frequently reported in asthmatic boys (Table 1). Although we cannot rule out that these results could have been influenced by the relatively small sample populations tested, the observed genotype differences between genders would not necessarily result only from sampling bias. However, in-breeding existing within the Saudi population could be a potential source of bias: the proportion of subjects married to close relatives was reported to range from 41% to 69%, depending on the region.32 Although we made efforts to recruit unrelated individuals, in practice it is difficult to obtain reliable information of the parental relationships. Importantly, the lack of deviation from the Hardy-Weinberg equilibrium suggested insignificant bias in our study due to inbreeding, stratification or sample selection. Gender is a known risk factor for asthma. For instance, asthma incidence can be strongly influenced by the distinct characteristic behavior of males and females and hence by other factors present in the environment in which they grow up.30,31 Nevertheless, at birth, twice as many males compared to females have been reported to have higher neonatal IgE levels, suggesting that gender-specific events have occurred in utero.33 Whether this is due to genetic or immunologic variation is still unclear. Also, some investigators have suggested that the airway size is smaller in boys when compared to girls of the same age, which may contribute to an increased risk of asthma progression, for example, after a viral infection.34
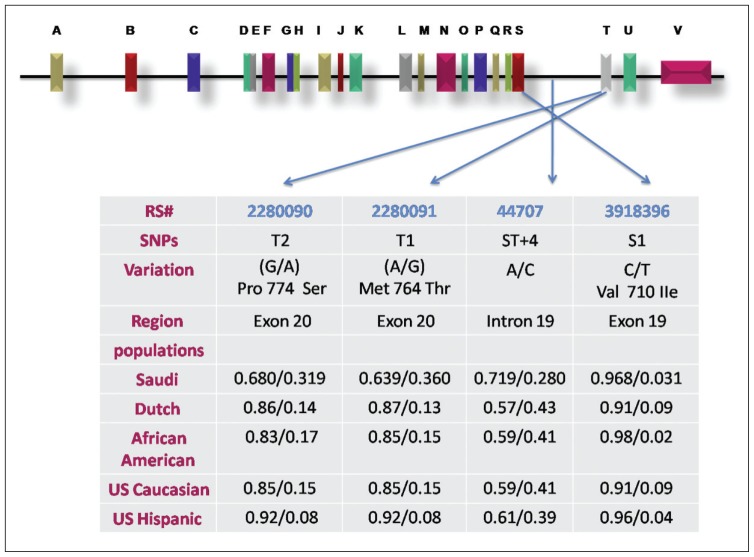
Our linkage disequilibrium analysis indicated that T1 and T2 were tightly associated, thus confirming the results of others.10 Moreover, this may explain the above observation, that male asthmatics have an increased risk of asthma because of the significantly higher frequency of T1 A/G and T2 G/A genotypes. Importantly, the high-frequency haplotype [G-A-A-C] encompassing T1 mutant G and T2 mutant A alleles, confirmed a significant association with the asthmatic phenotype. These observations suggest that Saudi children may have a genetic susceptibility to asthma. Arguably, the present haplotype analysis could be of limited reliability, based on the relatively small sample populations tested. However, this analysis could be a useful reference in future works employing larger sample populations in Saudi Arabia, and could be compared to that of other ethnic groups. In this regard, a comparative meta-analysis of allele frequencies for T1, T2, ST+4 and S1 using the present data from the Saudi children showed that the T2 mutant A allele and the T1 mutant G allele occurred significantly at much higher frequencies than in other asthmatic ethnic groups (Dutch, African American, United States whites and Hispanics) (Figure 1). Moreover, the results from the Saudi population are also in agreement with those observed in United States whites and Hispanics, showing a significant association between the T2 G/A polymorphism and asthma. The T1 mutant G allele was also found to be significantly associated with asthma development, as reported in a meta-analysis study involving 8 different ethnicities.35 In conclusion, the ADAM33 mutant alleles and genotypes for T2 and T1 SNPs, but neither ST+4 nor S1, were found to be associated significantly with asthma development in Saudi asthmatic children. Reports of inconsistent results among distinct ethnic groups may suggest that not all the critical haplotypes for asthma susceptibility in ADAM33 have been unambiguously identified. These results require further replication in larger sample population studies. We are currently recruiting more patients and subjects and expect to get up to 2000 patients and at least an equal number of healthy controls, to confirm which haplotypes are associated with increased risk of asthma in the Saudi population.
Figure 1.
Genomic structure of ADAM33 and the location of the 4 SNPs genotype. The 22 exons are shown as boxes and designated by the letters A through V. The SNPs and allele frequencies in Saudi compared to other populations are shown in the table.
REFERENCES
- 1.National Heart Lung and Blood Institute. NHLI/WHO Workshop report. 1995. Global initiative for asthma. Global strategy for asthma management and prevention. [Google Scholar]
- 2.Koopman LP, Brunekreef B, DeJongste JC, Neijens HJ. Definition of respiratory symptoms and disease in early childhood in large prospective birth cohort studies that predict the development of asthma. Pediatric Allergy and Immunol. 2001;12:118–124. doi: 10.1034/j.1399-3038.2001.012003118.x. [DOI] [PubMed] [Google Scholar]
- 3.Grant EN, Wagner R, Weiss KB. Observations on emerging patterns of asthma in our society. J Allergy Clin Immunol. 1999;104:S1–S9. doi: 10.1016/s0091-6749(99)70268-x. [DOI] [PubMed] [Google Scholar]
- 4.The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee. Worldwide variation in prevalence of symptoms of asthma, allergic rhino-conjunctivitis, and atopic eczema: ISAAC. Lancet. 2001;27:313–314. [Google Scholar]
- 5.Al-Ghamdi BR, Mahfouz AA, Abdelmoneim I, Khan MY, Daffallah AA. Socioclinical profile of children with asthma in Al-Majmaah Health Province. Saudi Medical Journal. 2000;21:847–851. [PubMed] [Google Scholar]
- 6.Al-Frayh AR, Shakoor Z, Gad elrab MO, Hasnain SM. Increased prevalence of asthma in Saudi Arabia. Ann Allergy Asthma Immunol. 2001;86:292–296. doi: 10.1016/s1081-1206(10)63301-7. [DOI] [PubMed] [Google Scholar]
- 7.John WS, Stephen SR, Larry B. Genetics of allergic disease. Allergy Clin Immunol. 2008;121:384–387. [Google Scholar]
- 8.Scott TW, Benjamin AR, Angela R. Asthma genetics and genomics 2009. Current Opinion in Genetics & Development. 2009;19:279–282. doi: 10.1016/j.gde.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 10.Schedel M, Depner M, Schoen C, Weiland SK, Vogelberg C, Niggemann B, et al. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respiratory Research. 2006;7:91. doi: 10.1186/1465-9921-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 12.Gao PS, Huang SK. Genetic aspects of asthma. Panminerva Med. 2004;46:121–134. [PubMed] [Google Scholar]
- 13.Dongju S, Ximei Z, Hong S, Fuzhen L, Lianhong J, Jing Z. Association of ADAM33 gene polymorphisms with adult allergic asthma and rhinitis in a Chinese Han population. BMC Medical Genetics. 2008;9:82. doi: 10.1186/1471-2350-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, et al. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clinical and Experimental Allergy. 2004;34:757–760. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- 15.Bijanzadeh M, Ramachandra NB, Mahesh PA, Savitha MR, Kumar P, Manjunath BS, et al. Association of IL-4 and ADAM33 Gene Polymorphisms with Asthma in an Indian Population. Lung. 2010;188:415–422. doi: 10.1007/s00408-010-9247-2. [DOI] [PubMed] [Google Scholar]
- 16.Matsusue A, Kiyohara C, Tanaka K, Sasaki S, Miyake Y. ADAM33 genetic polymorphisms and risk of atopic dermatitis among Japanese children. Clin Biochem. 2009;42:477–483. doi: 10.1016/j.clinbiochem.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Hirota T, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, et al. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin Exp Allergy. 2006;36:884–891. doi: 10.1111/j.1365-2222.2006.02522.x. [DOI] [PubMed] [Google Scholar]
- 18.Thongngarm T, Jameekornrak A, Limwongse C, Sangasapaviliya A, Jirapongsananuruk O, Assawamakin A, et al. Association between ADAM33 Polymorphisms and Asthma in a Thai Population. Asian Pacific Journal of Allergy and Immunology. 2008;26:205–211. [PubMed] [Google Scholar]
- 19.Vergara CI, Acevedo N, Jiménez S, Martínez B, Mercado D, Gusmão L, et al. Six-SNP haplotype of ADAM33 is associated with asthma in a population of Cartagena, Colombia. Int Arch Allergy Immunol. 2010;152:32–40. doi: 10.1159/000260081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Undarmaa S, Mashimo Y, Hattori S, Shimojo N, Fujita K, Miyatake A, et al. Replication of genetic association studies in asthma and related phenotypes. Human Genet. 2010;55:342–349. doi: 10.1038/jhg.2010.32. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Liu QJ, Li JS, Li HC, Wei CH, Guo CH, Gong YQ. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet. 2006;33:303–306. doi: 10.1111/j.1744-313X.2006.00617.x. [DOI] [PubMed] [Google Scholar]
- 22.Lind DL, Choudhry S, Ung N, Ziv E, Avila PC, et al. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med. 2003;168:1312–1316. doi: 10.1164/rccm.200306-877OC. [DOI] [PubMed] [Google Scholar]
- 23.Shu Y, Dhillon S, Ke X, Collins AR, Day INM. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:e88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNP Stats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 25.Haagerup A, Bjerke T, Schiqtz PO, Binderup HG, Dahl R, Kruse TA. Asthma and atopy – a total genome scan for susceptibility genes. Allergy. 2002;57:680–666. doi: 10.1034/j.1398-9995.2002.23523.x. [DOI] [PubMed] [Google Scholar]
- 26.Howard TD, Meyers DA, Bleecker ER. Mapping susceptibility genes for asthma and allergy. J Allergy Clin Immunol. 2000;105:477–481. doi: 10.1016/s0091-6749(00)90046-0. [DOI] [PubMed] [Google Scholar]
- 27.Jie Z, Hu Z, Bai C, Jin M. ADAM33 Gene Polymorphisms Associate with Asthma Susceptibility and Severity in East China Han Population. J Asthma. 2011;48:979–85. doi: 10.3109/02770903.2011.624233. [DOI] [PubMed] [Google Scholar]
- 28.Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, et al. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112:717–722. doi: 10.1016/s0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- 29.Simpson A, Maniatis N, Jury F, Cakebread JA, Lowe LA, Holgate ST, et al. Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33) predict impaired early-life lung function. American J Respir Critical Care Med. 2005;172:55–60. doi: 10.1164/rccm.200412-1708OC. [DOI] [PubMed] [Google Scholar]
- 30.Anderson HR, Pottier AC, Strachan DP. Asthma from birth to age 23: incidence and relation to prior and concurrent atopic disease. Thorax. 1992;47:537–542. doi: 10.1136/thx.47.7.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sears MR, Burrows B, Flannery EM, Herbison GP, Holdaway MD. Atopy in childhood. I. Gender and allergen related risks for development of hay fever and asthma. Clin Exp Allergy. 1993;23:941–948. doi: 10.1111/j.1365-2222.1993.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 32.Al Husain M, Al Bunyan M. Consanguineous marriages in a Saudi population and the effect of inbreeding on prenatal and postnatal mortality. Ann Trop Paediatr. 1997;17:155–60. doi: 10.1080/02724936.1997.11747879. [DOI] [PubMed] [Google Scholar]
- 33.Kerkhof M, Wijga A, Smit HA, de Jongste JC, Aalberse RC, Brunekreef B, et al. The effect of prenatal exposure on total IgE at birth and sensitization at twelve months and four years of age: The PIAMA study. Pediatric Allergy and Immunology. 2005;16:10–18. doi: 10.1111/j.1399-3038.2005.00217.x. [DOI] [PubMed] [Google Scholar]
- 34.Becklake MR, Kauffmann F. Gender differences in airway behaviour over the human life span. Thorax. 1999;54:1119–38. doi: 10.1136/thx.54.12.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blakey J, Halapi E, Bjornsdottir US, Wheatley A, Kristinsson S, Upmanyu R, et al. Contribution of ADAM33 polymorphisms to the population risk of asthma. Thorax. 2005;60:274–276. doi: 10.1136/thx.2004.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]