Figure 5.
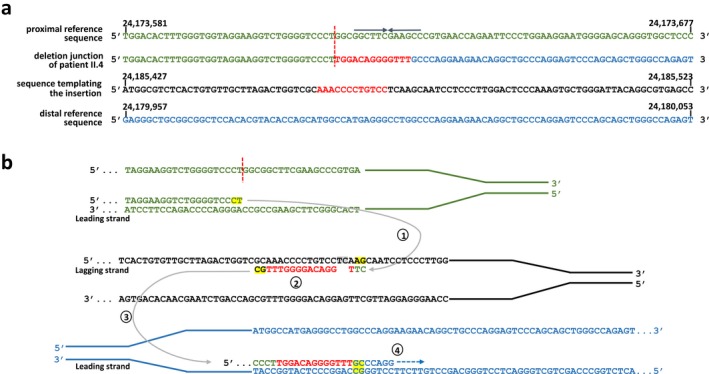
The germline SMARCB1 deletion investigated in this study, as well as the insertion of 13‐bp (red) at the deletion breakpoints, most likely resulted from replication‐associated template switching. (a) Alignment of the deletion breakpoint‐flanking sequences of patient II.4 against the reference sequence of the human genome (hg19). Sequences located at the proximal (centromeric) deletion breakpoint are indicated in green, while sequences at the distal (telomeric) breakpoint are given in blue. The vertical red line highlights the position of the proximal deletion breakpoint. The 13‐bp insertion (red) identified at the deletion junction exhibits homology to a sequence located 5.5‐kb telomeric to the distal breakpoint region. (b) Model proposed to explain the origin of the deletion‐associated insertion. In the proximal breakpoint‐flanking region, DNA synthesis at the leading strand is interrupted but appears to have resumed, after an interstrand template switch, to a replication fork located 5.5‐kb telomeric to the distal breakpoint‐flanking region (step 1). Subsequently, the 13‐bp indicated in red are newly synthesized and included in the nascent DNA strand at the replication fork (step 2). Single‐nucleotide changes due to DNA polymerase errors are highlighted in gray. Subsequently, another template switch occurs on the leading strand (step 3) upon which replication is continued (step 4). The nucleotides exhibiting microhomology at sites of template switching are marked in yellow. An inverted repeat of 6‐bp marked by arrows was identified close to the proximal deletion breakpoint which may have caused a cruciform structure responsible for replication stalling
