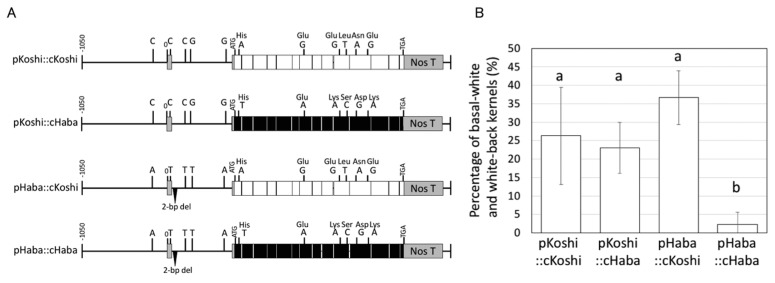
Fig. 5.
Promoter-swap analysis of Sus3. (A)Vector constructs for promoter-swap analysis. Promoter sequences of Sus3Koshi (pKoshi) and Sus3Haba (pHaba) were fused to the coding cDNA sequences of Sus3Koshi (cKoshi) and Sus3Haba (cHaba). All four constructs were fused with the Nos terminator at the end of the coding sequence. (B) High-temperature treatment test of promoter-swap plants. The four constructs indicated in (A) were transformed into ‘Nipponbare’. Transformants were grown under high-temperature condition during the ripening stage. The percentages of basal-white and white-back kernels were measured to compare high-temperature tolerance. Error bars indicate standard deviations. Different letters above the bars indicate a significant difference at 5% (Tukey’s HSD test), n > 3.