Figure 5.
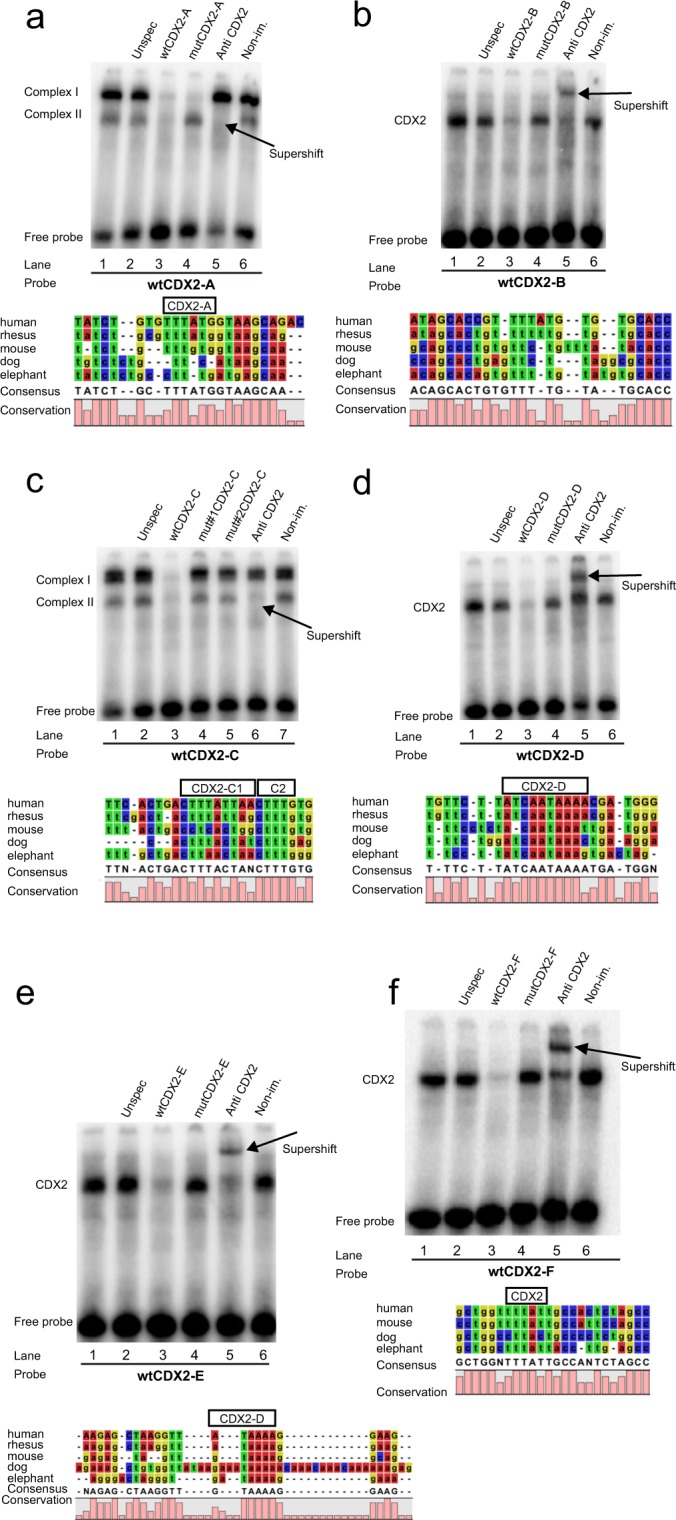
CDX2 binds all the predicted binding-sites within the ST14 and SPINT1 enhancers in Caco-2 cells in vitro. Gel shift analysis of double-stranded [γ32]ATP-labeled oligonucleotides wtCDX2-A (a) wtCDX2-B (b) wtCDX2-C (c) wtCDX2-D (d) wtCDX2-E (e) covering the putative CDX2 binding sites within the ST14 enhancer and one within the SPINT1 enhancer (f) [γ32] ATP-labeled probes were incubated with Caco-2 nuclear cell extracts, and all resulted in shifted DNA/protein complexes, depicting that binding has taken place (lane 1). Competition by excess of unlabeled unspecific oligonucleotides (lane 2), unlabeled wild-type oligonucleotides (wt) (lane 3) and unlabeled mutated oligonucleotides (mut) (lane 4 (for C lanes 4 and 5)), were used for validating the specificity of the shifted DNA/protein complexes. Addition of anti-human CDX2 antibody was used for the supershift assays (lane 5 (for C lane 6)), and non-immune serum was used as negative control (lane 6 (for C lane 7)). Supershifted complexes are indicated by red arrows. Gels are representative of three independent EMSA assay experiments.
