Fig. 4.
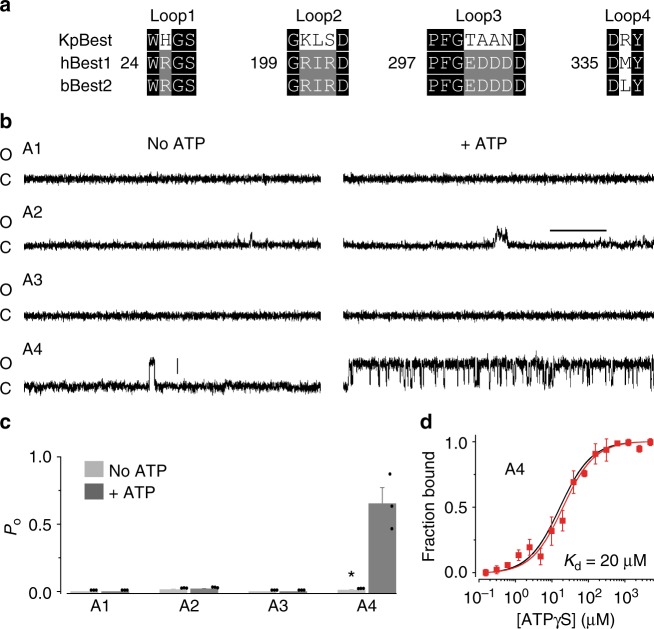
Mapping ATP-binding motif(s) in KpBest. a Candidate ATP-binding motifs in KpBest, hBest1, and bBest2. Black background, identical residues in all three sequences; gray background, identical residues in two sequences. Numbers indicate the position of the first residue in each motif on hBest1. b Current traces of single KpBest mutant channels recorded from planar lipid bilayers at 80 mV in the absence or presence of 2 mM ATP (Scale bar, 3.5 pA, 250 ms). c Bar chart showing the open probability of KpBest mutant channels in the absence or presence of 2 mM ATP. n = 3 for each bar. *P < 0.05 compared to currents from the same channels in the presence of ATP, using two-tailed unpaired Student t test. d The MST binding curve of KpBest A4 to ATPγS (red). Protein fraction bound vs. [ATPγS], n = 3 for each point. The binding curve of WT KpBest to ATPγS (black) is shown for comparison. All error bars in this figure represent s.e.m.