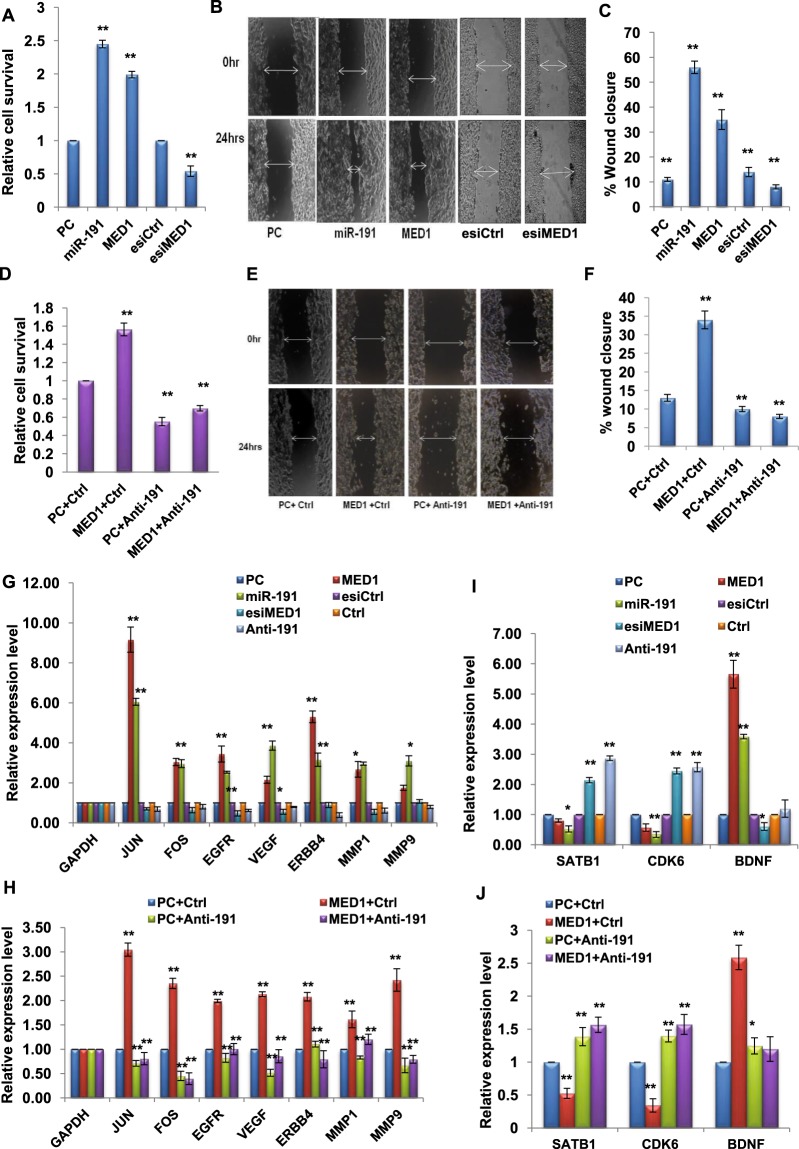
Figure 6.
miR-191 acts as the downstream effecter of MED1 mediated cellular functions. (A–C) Functional implications of MED1 or miR-191 overexpression in breast cancer. MCF7 cells were transfected with MED1 or miR-191 or PC (control vector) and MTT assay was performed (48 hrs post transfection) to measure effect on cell survival. The results show that both MED1 and miR-191 promote cell survival (A). (B,C) Wound healing assay was performed to observe the effect of miR-191 and MED1 on cell migration. MCF7 cells were transfected with MED1 or miR-191 or control vector and gap or wound closure was observed (B) using Nikon microscope, 10X magnification and % gap closure was quantified using arbitrary scale (C). Graph shows that more healing (gap closure) was observed in response to both miR-191 and MED1 overexpression, hence both MED1 and miR-191 impart enhanced cell migratory capacity to the cells. (D–F) MED1 mediated cellular effects are miR-191 dependent. MCF7 cells were transiently transfected with MED1/PC (vector control) along with inhibition of miR-191(anti-miR-191)/Ctrl and effect on cell survival was looked for using MTT assay (D). Increase in cell survival due to MED1 overexpression was significantly reduced when miR-191 was inhibited along with MED1 overexpression (E). Wound healing assay was performed to confirm the effect of MED1 and miR-191 on cell migration. Levels of MED1 were transiently overexpressed along with/without miR-191 inhibition (using anti-miR-191/ctrl) and gap closure was observed after 24 hrs. Comparatively, lesser gap was filled when MED1 overexpression was coupled with miR-191 inhibition (F) when quantified using arbitrary scale thereby, confirming that miR-191 is a downstream effecter for MED1 mediated cellular migration. (G) qRT-PCR data showing transcript levels of JUN, FOS, EGFR, VEGF, MMP1 and ERBB4 on miR-191 or MED1 overexpression/inhibition. GAPDH has been used for normalization of qRT-PCR data. (H) qRT-PCR data showing transcript levels of JUN, FOS, EGFR, VEGF, MMP1 and ERBB4 on MED1 overexpression or miR-191 inhibition. (I) qRT-PCR data showing transcript levels of established miR-191-target genes (SATB1, CDK6 and BDNF) in response to both miR-191 or MED1 overexpression/inhibition. GAPDH has been used for normalization of qRT-PCR data. (J) qRT-PCR data showing transcript levels of established miR-191-target genes (SATB1, CDK6 and BDNF) on MED1 overexpression or miR-191 inhibition. (**P<0.05, (**P<0.05, * P<0.1). Error bars denote + SD.