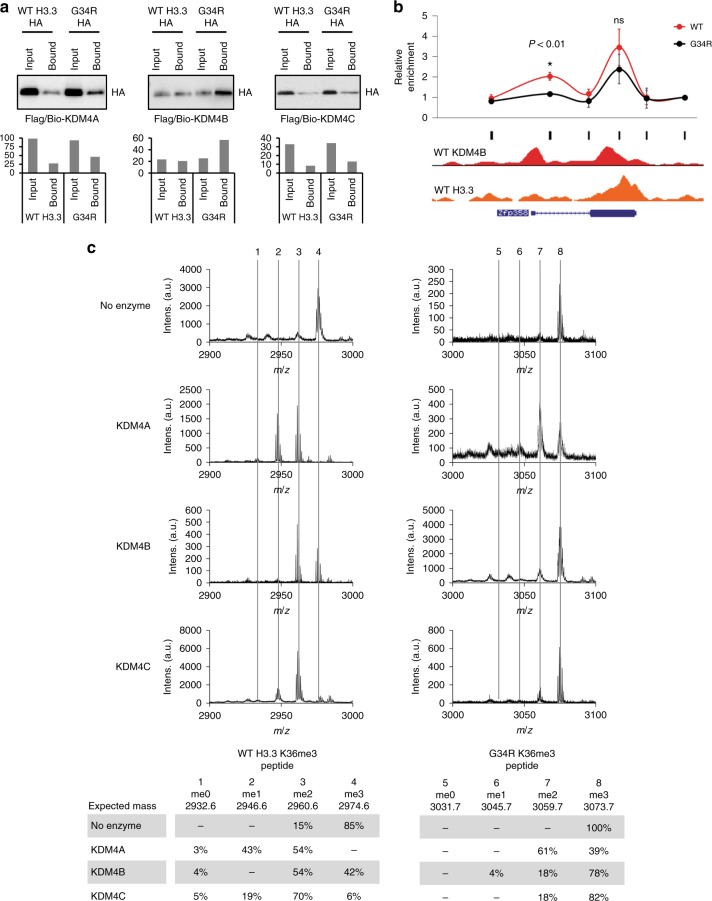
Fig. 4.
H3.3 G34R preferentially binds to KDM4 and inhibits its demethylase activity. a Protein pulldowns of KDM4-A, -B and -C, immunoblotted with anti-HA for detection of HA-tagged WT H3.3 and H3.3 G34R. Representative blots are shown and quantitation values are shown. b ChIP-qPCR of KDM4B in WT and G34R cells tiled across a representative gene, Zfp358. Results are normalised for input and values represent mean enrichment of three independent experiments, calculated relative to a negative control (Zfp358, primers 6). Data are mean average and error bars are standard deviations of three independent experiments (n = 3). P-values calculated using Student’s T-test (*P < 0.01). Black bars indicate primer locations. ChIP-seq profiles of KDM4B and H3.3 in WT cells across the gene are shown. c Mass spectrometry analysis of in vitro demethylase assays. KDM4-A, -B and -C were combined with either WT H3.3 K36me3 peptide (left panel) or a G34R K36me3 peptide (right panel). Dashed lines indicate expected masses for K36 me0, me1, me2 and me3