Abstract
In recent years, thyroid malignances have become more prevalent, especially among women. The most common sporadic types of thyroid tumors of follicular origin include papillary, follicular and anaplastic thyroid carcinomas. Although modern diagnosis methods enable the identification of tumors of small diameter, tumor subtype differentiation, which is imperative for the correct choice of treatment, is still troublesome. This review discusses the recent advances in the field of molecular marker identification via next-generation sequencing and microarrays. The potential use of these biomarkers to distinguish among the most commonly occurring sporadic thyroid cancers is presented and compared. Geographical heterogeneity might be a differentiator, although not necessarily a limiting factor, in biomarker selection. The available data advocate for a subset of mutations common for the three subtypes as well as mutations that are unique for a particular tumor subtype. Tumor heterogeneity, a known issue occurring within solid malignancies, is also discussed where applicable. Public databases with datasets derived from high-throughput experiments are a valuable source of information that aid biomarker research in general, including the identification of molecular hallmarks of thyroid cancer.
Keywords: Thyroid cancer, Biomarkers, NGS, Molecular markers, PTC, FTC, ATC
Background
Thyroid cancer (TC) is one of the most frequent endocrine malignancies, accounting for 3–4% of cancers [1], and its occurrence has increased by approximately 5% on a yearly basis, with higher prevalence in females than in males (20.6 vs. 6.9 new cases per 1000 persons) [2]. The number of newly diagnosed cases has risen dramatically in the last 10 years, which could be partially ascribed to the availability of more sensitive diagnostic tools, i.e., ultrasonography and fine-needle aspiration (FNA) and the smaller size of diagnosed tumors. However, over diagnosis is also an issue because its occurrence rate has risen 15-fold since 2003, whereas mortality rates have not changed [3].
In general, the 5- and 10-year survival rates for TC patients are excellent (approx. 98%) but are related to the age of the patient at the time of diagnosis and the cancer subtype [1, 4, 5].
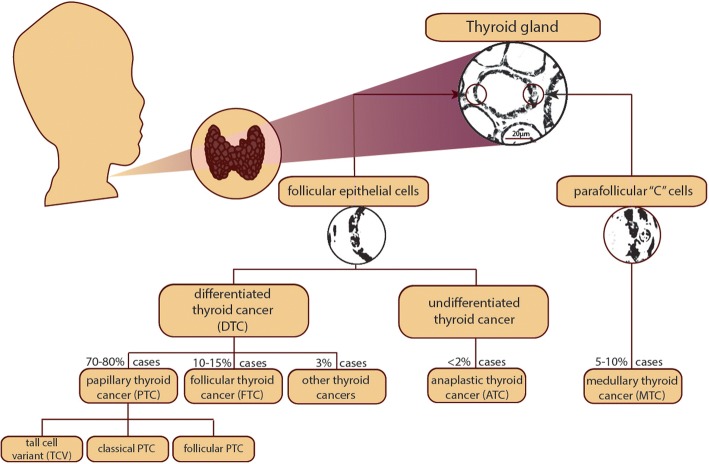
Both papillary (PTC) and follicular thyroid carcinoma (FTC) arise from follicular epithelial thyroid cells involved in iodine metabolism. PTC and FTC, together with the less common Hürtle cell carcinoma, are classified as differentiated thyroid cancer (DTC, see Fig. 1) [6, 7]. Both PTC and FTC progress slowly and are generally characterized by good prognosis, especially if diagnosed early [5].
Fig. 1.
Overview of thyroid cancer types and their origins
Undifferentiated anaplastic thyroid carcinoma (ATC) is the most aggressive TC type. Although ATC also originates from follicular cells, similar to PTC and FTC, it does not possess their original biological properties [8]. ATC represents 2–5% of cases, (77% in women) with the worst prognosis and a 5-year survival rate of 5% [3]. ATC is insensitive to conventional methods of treatment [9].
In contrast, medullary thyroid cancer (MTC) is derived from parafollicular thyroid “C” cells, which produce calcitonin [2].
The majority of TC cases are sporadic, with only 5% of DTC characterized as familial (mostly PTC) and ~ 25% of MTC inherited as an autosomal trait [10]. Only sporadic tumors are analyzed in this review.
Although most mutations found in TC differ among types, certain DNA alterations were found to be common in more than one subtype. As discussed later in this review, ATC tumors appear to derive from other differentiated tumors and thus possess a large overlap with mutations present in DTCs, such as TMPRSS4. Mutations in certain genes, e.g., CHEK2, are reported in both PTC and FTC, although not with the same prevalence [11, 12], and their potential contribution to TC carcinogenesis is described in the respective paragraphs. In this work, we focus on tumor heterogeneity and the mutation burden carried by thyroid tumors, as tested primarily by high-throughput methods performed within larger genomic projects, including The Cancer Genome Atlas (TCGA).
We gathered the data from RNA expression and DNA sequencing experiments and identified potential genetic biomarkers of disease progression. Genome-wide association studies (GWAS) as well as sequencing and microarrays were considered. In this work, we present an overview of the available biomarkers candidates for progression and development of thyroid cancer and drivers of carcinogenesis, as discussed in detail in the respective sections. All gene functions were inferred using GeneCards (www.genecards.org) [13].
Genome-wide studies significantly aid in the identification of cancer-specific germline and somatic mutations, which can contribute to more sensitive diversification of cancer subtypes and facilitate early diagnosis. Identification of disease-specific point mutations can accelerate the evaluation of candidate target genes for therapeutic drugs and the search for novel driver mutations. However, the identification of polymorphisms (SNPs) could additionally improve prognosis and patient outcomes.
Common genetic determinants of thyroid cancer subtypes
In recent years, the development of sequencing and microarray technologies has permitted a whole-genome search for TC-linked or associated genes. Genome-wide association studies (GWAS) are a highly potent method for identification of high-incidence single nucleotide polymorphisms (SNPs) and copy number variations (CNVs). Recently, GWAS were used to study large TC patient cohorts [14–17] and were followed by studies confirming the findings [18–27]. Mutation hot spots identified through GWAS (microarray, next-generation sequencing (NGS) and high-resolution melting (HRM)) are collected in Table 1. Specific SNPs could be associated with susceptibility to DTC (mostly papillary and follicular) in single or multiple populations with variable strength.
Table 1.
Somatic mutations associated with susceptibility to differentiated thyroid cancers
| Chromosomal location | DbSNP identification No. | Gene | Gene function | Cancer type | Tested population | Literature |
|---|---|---|---|---|---|---|
| 1p31.3 | rs334725 | NFIA | Nuclear transcription factor | PTC, FTC | Icelandic, American, Dutch, Spanish | Gudmundsson et al., 2012 |
| 1q42.2 | rs12129938 | PCNXL2 | Correlated with tumorigenesis of colorectal carcinomas | DTC | Icelandic, American, Spanish, Dutch | Gudmundsson et al., 2017 |
| 2q35 | rs966423, rs6759952 | DIRC3 | lincRNA | PTC, FTC | Icelandic, American, Dutch, Spanish, Polish | Gudmundsson et al., 2012, Liyanarachchi et al., 2013 |
| DTC | Italian, Polish, Spanish, English | Köhler et al., 2013 | ||||
| 3q25.32 | rs7617304 | RARRES1 | Membrane protein gene responsive to retinoid acid | DTC | Italian | Köhler et al., 2013 |
| 3q26.2 | rs6793295 | LRCC34 near TERC (missense) | RNA telomerase | PTC, FTC | Icelandic, American, Spanish, Dutch | Gudmundsson et al., 2017 |
| 4q34.3 | rs17739370 TT variant | NEIL3 | DNA repair, BER | DTC | Italian | Cipollini et al., 2016 |
| 5q22.1 | rs73227498 | NREP and EPB41L4A | Intergenic region | PTC, FTC | Icelandic, American, Spanish, Dutch | Gudmundsson et al., 2017 |
| 5 | rs13184587 | ARSB intron | Intron of lysosomal sulfatase | DTC | Italian | Figlioli et al., 2014 |
| 7q21 | rs10238549, rs7800391 | IMMP2L | Processing of signal peptides in mitochondrial membrane | DTC | Italian | Köhler et al., 2013 |
| 8p12 | rs2439302 | NRG1 | membrane glycoprotein, signaling mediator | PTC, FTC | Icelandic, American, Dutch, Spanish | Gudmundsson et al., 2012 |
| 8q24 | rs6983267 | ncRNA | N/A | PTC | English | Jones et al., 2012 |
| 9q3.3 | rs10781500 | SNAPC4 | Large subunit of the DNAP complex | DTC | Italian | Köhler et al., 2013 |
| 9q22.33 | rs965513, rs1867277 (5’UTR region), rs71369530 | Proximity to FOXE1 | Deregulation of thyroid morphogenesis | PTC | Icelandic, Caucasian, Asian, Cuban, English, Belarussian, French Polynesian | Jones et al., 2012, Gudmundsson et al., 2009, Liyanarachchi et al., 2013, Damiola et al., 2014, Wang et al., 2016, Pereda et al., 2015, Maillard et al., 2015 |
| 10q24.33 | rs7902587 | near OBFC1 | Stimulator of DNA replication initiation factor | PTC, FTC | Icelandic, American, Spanish, Dutch | Gudmundsson et al., 2017 |
| 11 | rs1801516 | ATM | Cell-cycle checkpoint, response to DNA damage | DTC | Cuban women after multiple pregnancies, French Polynesian | Pereda et al., 2015, Maillard et al., 2015 |
| 13 | rs1220597 | SPATA13 intron | Regulation of cell migration and adhesion, guanine nucleotide exchange factor | DTC | Italian | Figlioli et al., 2014 |
| 14q13.3 | rs116909374 | NKX2–1 | Thyroid-specific transcription factor | PTC, FTC | Icelandic, American, Dutch, Spanish, Polish | Gudmundsson et al., 2012, Liyanarachchi et al., 2013 |
| 14q13.3 | rs944289 | Close to NKX2–1 | Thyroid-specific transcription factor | PTC, FTC | Icelandic, Cuban, English, American, Polish, French Polynesian | Jones et al., 2012, Gudmundsson et al., 2009, Liyanarachchi et al., 2013, Pereda et al., 2015, Maillard et al., 2015 |
| 14 | 241(Thr > Met) | XRCC3 | DNA repair, homologous recombination | DTC | Chinese, Iranian, Caucasian Portuguese | Wang et al., 2015, Fayaz et al., 2014, Bastos et al., 2009 |
| 14 | rs10136427 | BATF | Transcription factor, negative regulator of AP-1/ATF transcriptional events | DTC | Italian, Polish, Spanish | Figlioli et al., 2014 |
| 15q22.33 | rs2289261, rs56062135 | SMAD3 | Transcriptional modulator | PTC, FTC | Icelandic, American, Spanish, Dutch | Gudmundsson et al., 2017 |
| 20 | rs7267944 | DHX35 | RNA helicases | DTC | Italian, Polish, Spanish | Figlioli et al., 2014 |
Variants determined by GWAS. DTC Unspecified differentiated thyroid cancer, PTC Papillary thyroid cancer, FTC Follicular thyroid cancer
Sixteen case/control studies allowed identification of 27 SNPs located primarily within the coding regions (see Table 1). Only rs6983267 was located in the non-coding RNA; rs1220597, rs73227498, and rs13184587 were located in the introns; and rs965513, rs1867277, rs71369530 and rs944289 were located in proximity to NKX2–1. This observation might stem from the fact that most microarray and NGS experiments are focused on transcriptome analysis and can be biased against regulatory or non-coding fragments. TC-associated genes are often connected to DNA-damage repair or transcription.
Using a slightly different approach, Gudmundsson et al., selected 22 SNPs based on a score of high association with high levels of thyroid stimulating hormone in a GWAS study of over 27,000 samples from an Icelandic population [16]. The results of genotyping of 561 samples of the non-medullary type were compared with over 40,000 controls from different populations (Dutch, American and Spanish). Three variants proved to be significantly correlated, namely, rs966423 in non-coding RNA-DIRC3 (OR = 1.34, P = 1.3·10− 9), rs2439302 membrane glycoprotein involved in cell signaling NRG1 (OR = 1.36, P = 2.0·10− 9) and rs116909374 in thyroid-specific transcription factor NKX2–1 (OR = 2.09, P = 4.6·10− 11), and their functions in thyroid tumorigenesis are still unknown.
The ThyroSeq microarray panel (ThyroSeq) is widely used and offers the possibility of testing more than 1000 hotspots in 14 TC-related genes and over 40 fusions simultaneously. Nikiforova and Nikiforov tested over 800 TC samples of all types using ThyroSeq panels, thus proving its usefulness in detection and classification of cancerous tissue [28–30].
Figlioli et al., performed SNP genotyping of an Italian population (case/controls: 1437/1534), validated in DTC patients from Poland (case/controls: 448/424) and Spain (case/controls: 375/408) [14]. The strongest correlation among all tested cohorts was found for rs10136427 localized in transcription regulator BATF, (OR = 1.40, P = 4.35·10− 7) and rs7267944 in putative RNA helicase DHX3 (OR = 1.39, P = 2.13·10− 8).
Gudmundsson et al., published a follow-up study in Icelandic, Dutch, Spanish and 2 American populations (case/controls: 1003/278,991, 85/4956, 83/1612, 1580/1628 and 250/363, respectively) confirming 5 novel loci associated with non-medullary thyroid cancer (Pcombined < 3 × 10− 8), i.e., rs12129938, rs6793295, rs73227498, and two independently associated variants, i.e., rs2289261 (OR = 1.23; P = 3.1·10–9) and rs56062135 (OR = 1.24; P = 4.9·10− 9) [31].
Applying a presumption that the DNA repair genes of base (BER) or nucleotide (NER) excision repair pathways might be involved in TC tumorigenesis, Cipollini et al., genotyped known SNPs in 450 case-control paired DTC samples from an Italian population [32]. The TT variant of base excision repair gene NEIL3, which codes a DNA glycosylase, was associated with increased risk of DTC. Another GWAS study on an Italian population performed by Köhler et al. associated mutations in non-coding RNA genes DIRC3, RARRES1, SNAPC4 and IMMP2L with increased DTC in a high-incidence population of 690 cases and 497 controls and confirmed this finding in 3 low-incidence populations (total of 2958 cases and 3727 controls) [15] (See Table 1). SNAPC4 encodes a large subunit of the RNA-activation protein complex, and RARRES1 and IMMP2L are transmembrane proteins.
Papillary thyroid Cancer (PTC)
Derived from follicular cells, papillary thyroid cancer is named after its cyto-architecture and can be further divided into 3 subtypes based on histotype: tall cell variant (TCV), follicular, and classical (most common) [33]. According to TCGA, up to 70% of somatic PTC drivers are found in activators of the MAPK pathway and include BRAF, RAS and rearrangements of the RET and NTRK1 genes [5] (See Table 2). The alterations are generally thought to be mutually exclusive in PTC [34–37], but contradictory data have emerged [38–41]. Other mutations such as PTEN and PIK3CA [42] have been reported at lower frequencies (2/86 (2.32%) and 3/86 (3.48%), respectively). The mutation density is relatively low at 0.41 mutations/Mb for PTC and 0.5 mutations/Mb for TCV. PTC is often multifocal, with a main tumor (> 1 cmØ) and several microcarcinomas [43, 44]. Nodules might be positioned unilaterally or bilaterally in the thyroid lobes. Multifocality is a characteristic of up to 40% of all PTC, [45, 46] leading to aggressiveness and resistance to radioiodine treatment [47]. The clonal origin of each singular carcinoma is not necessarily the same because tumors might arise independently through a series of molecular events, such as chromosome X inactivation [43, 48–52], but certain authors suggest clonal homogeneity between the nodules [49, 53–57].
Table 2.
Somatic mutations characteristic of PTCs
| Gene | Localization | Gene function | Mutation | Defect in cancer | Clinical correlation | Literature |
|---|---|---|---|---|---|---|
| Gene | ||||||
| BRAF | 7q34 | Serine/threonine kinase, response to cell growth factors | B-type Raf Kinase (chr 7) 2 Missense mutation V600E (T < A 1799), rs113488022 | Constitutive activation of MAPK pathway | Positive correlation with age, marker of TCV subtype | TCGA, Kimbrell et al., 2015, Lu et al., 2016, Gandolfi et al., 2013, Kim et al., 2006, Guerra et al., 2012, Sun et al., 2016, Gertz et al., 2016, Iyer et al., 2015, Lee et al., 2016 |
| CHEK2 | 22q12.1 | Cell cycle checkpoint kinase | IVS2 + 1G > A, 1100delC or del5395, missense mutation I157T | DNA repair mechanism dysfunctions | Positive correlation with cancer aggressiveness | Siolek et al., 2015, Wójcicka et al., 2014, Kaczmarek-Ryś et al., 2015 |
| DLL4 | 15q15.1 | Notch signaling mediator | Patient specific mutations | Promotes angiogenesis | Correlated with presence of lymph node metastases | Le Pennec et al., 2015 |
| EIF1AX | Xp22.12 | Translation initiation factor, transfer of Met-tRNAf | Hotspot at A113_splice site intron 5/exon 6 | Potential driver mutation | N/A | TCGA, Forbes 2011, Martin 2013, Karanamurthy 2016 |
| FOXE1 | 9q22.33 | Transcription factor | rs965513 AA, AG; rs944289; c.821C > A, p.P54Q; c.943A > C p.K95Q; c.994C > T, p.L112F | Deregulation of thyroid morphogenesis | Thyroid cancer susceptibility marker | Mond et al., 2015, Gudmundsson et al., 2009, Penna-Martinez et al., 2014 |
| PIK3CA | 3q26.32 | PI3K/AKT/mTOR pathway effector | E545A | Mutation of helical domain | N/A | Lee et al., 2016 |
| PTEN | 10q23.31 | PI3K/AKT/mTOR pathway effector | N/A | Produces a truncated protein | N/A | Xing et al., 2013 |
| RAS | 11p15.5, 1p13.2, 12p12.1 | Signal transduction | H-Ras (chr11), N-Ras (chr1), K-Ras (chr12) | Preferential activation of PI3K-AKT pathway | Positive correlation with cancer aggressiveness | Rossi et al., 2015, Gertz et al., 2016, Abubaker et al., 2008, Zou et al., 2014 |
| TERT promoter | 5p15.33 | Telomerase reverse transcriptase | C > T 1295228 and 1,295,250 C > A at 1295250 | Gain of immortality | Positive correlation with cancer aggressiveness | Bae et al., 2016, Liu et al., 2014, Liu et al., 2013, Sun et al., 2016 |
| Chromosomal Abberations | ||||||
| RET | 10q11.21 | Tyrosine kinase transmembrane receptor | Rearrangements: RET/PTC1, RET/PTC2, RET/PTC3, RET/PTC4. RET/PTC5, RET/PTC6, RET/PTC7, RET/PTC8, RET/PTC9, PCM1-RET, EKLS-RET, FKBP-RET, RET-ANK3, TBL1XR1-RET, AKAP13-RET, ERC1-RET, HOOK3-RET, SPECC1L-RET, ACBD5-RET, ΔRFP-RET | Downstream signaling of MAPK and PI3K pathways, evasion of apoptosis | Common in pediatric PTC, common co-occurrence with BRAF mutation | TCGA, Gertz et al., 2016, Rossi et al., 2015, Hamatani et al., 2014, Corvi et al., 2000, Ciampi et al., 2007, Klugbauer et al., 1998, Salassidis et al., 2000, Saenko et al., 2003, Nakata et al., 1999, Hamatani et al., 2014, Bongarzone et al., 1993, Grieco et al., 1990 |
| Abnormal expression | ||||||
| ATP5E | 20q13.32 | ATPase subunit 5E | Down-regulation | Disruption of ATP synthesis in mitochondria | Potential PTC biomarker | Hurtado-Lopez et al., 2015 |
| MUC1 | 1q22 | Proliferation and signaling of epithelial cells | Overexpression | Leads to propagation of tumorigenesis and metastasis | Poor outcome marker | Renaud et al., 2014 |
| TMPRSS4 | 11q23.3 | Serine protease | Overexpression | Migration and metastasis of cancer cells | Malignant tumors | Kebebew et al., 2005, Jarzab et al., 2005, Guan et al., 2015 |
| YY1 | 14q32.2 | Transcription factor | Overexpression | Leads to increased cell proliferation | Positive correlation with age | Arribas et al., 2015 |
| Regulation Of Expression | ||||||
| micro RNA | Xp11.3 | Regulation of expression of affiliated genes | let-7 miRNA overexpression | Disruption of regulatory pathways (e.g. DNA damage response, stress response), propagation of cancer growth and expansion through down/up-regulation of target genes | N/A | Salajegheh et al., 2016, Yoruker et al., 2016, Lee et al., 2013, Zhang et al., 2010, Lei et al., 2016, Hong et al., 2016, Samsonov et al., 2016, Hu et al., 2017 |
| 9p21.3 | miR-31 overexpression | |||||
| 8q24.3 | miR-146b overexpression | |||||
| 19q13.41 | miR-151-5p overexpression | |||||
| 10q24.32 | miR-221 overexpression | |||||
| Xp11.3 | miR-222 overexpression | |||||
| 17q23.1 | miR-21 down-regulation | |||||
| 9q34.3 | miR-126 down-regulation | |||||
| 19p13.12 | miR-20b | |||||
| Xq26.2 | miR-639 | |||||
Genetic alterations in kinases
BRAF
The most common somatic mutation occurring in PTC is a mis-sense BRAF mutation resulting in thymine-to-adenine substitution at position 1799 of the B-type Raf Kinase (BRAF) gene. This mutation leads to a valine-to-glutamate substitution at codon 600 of the BRAF protein (BRAFV600E) and constitutive activation of the MAPK signaling pathway via activation of the G-coupled receptor in the membrane [58–60], and it is common for several cancers, including non-small cell lung cancer and melanomas. [59–61]. BRAF is an activator of BRAF-activated non-coding RNA (BANCR), which regulates many cellular processes, including tumorigenesis, metastasis and, apoptosis [62]. BRAF can function as both a tumor suppressor and disease progression factor [63]. BRAFV600E is typical for TCV and classical subtypes, whereas RAS mutations predominantly drive the follicular subtype [33, 64]. This dependence, in combination with the various prevalence of driver mutations in populations, might explain certain of the disparities between different studies.
Recently, the potential heterogeneity of BRAF mutants (intra- and inter-tumoral) has been emphasized using both traditional methods (PCR verified by Sanger sequencing) as well as novel techniques such as exome capture and pyrosequencing. Kimbrell et al., tested 57 tumors from 27 patients for the presence of the BRAF V600E mutation [65]. The results were discordant between primary and secondary tumors in 10 out of 27 cases, but no significant histological changes were observed. However, the irregularity of the tumor edge appears to indicate its metastatic origin. No correlation was detected for the lobe positioning of the concordant and discordant nor the size of BRAF-positive and negative tumors. Sun et al., showed (n = 455) that 75.5% of the patients in a Chinese population harbored a BRAFV600E mutation, which was significantly correlated with increasing patient age [66]. In contrast, the rate of BRAFV600E mutations was two times lower in children than in adults [67]. One of 14 pediatric patient samples was positive for concomitant BRAF mutation and RET/PTC3 rearrangement (see below). Lu et al., identified BRAFV600E mutation as the most common using deep sequencing of 21 foci from 8 patients [68]. The experiments confirmed that multifocal TC could be heterogeneous and that BRAF is not necessarily the driver because up to 75% of the clones had independent clonal origins. Those results were supported by reports from other groups in which foci did not share the same mutation patterns [48, 69–71]. Gandolfi et al., tested 37 primary PTC tumors and 95 metastases in adults and found that 43.9% of the samples were BRAF-positive, but no correlation was observed with metastasis. The allele percentage shows that BRAF mutations are heterogeneous and rarely a result of a clonal event [69, 72]. De Biase et al., tested the distribution of neoplastic cells in BRAFV600E-positive tumors (n = 85/155) [51]. The percentage of cells harboring a mutated BRAF allele present in each sample varied from less than 30% (n = 9/85) to 80% (n = 39/85). Down-regulation of the transcript was observed in paired PTC tumor samples and normal adjacent tissues. Real-time PCR shows that the down-regulation of BANCR correlates with patient prognosis with consideration of tumor size, number of nodules, stage, gender, metastasis and extrathyroidal extensions but not with age.
PIK3CA
Mutations in PIK3CA, a catalytic subunit of the phosphatidylinositol 3-kinase and a component of the PI3K/Akt signaling pathway, were found by Lee et al. in a targeted sequencing experiment (n = 240). One sample carried a PIK3CAE542K mutation (0.4%), 24 p.E545A mutation (10%) and 138 concomitant BRAFV600E and PIK3CAE454A mutations (57.7%) [73]. Independently, Wang et al., found 20 samples carrying the PIK3CA copy gain mutation (14%, n = 141) [74].
RET proto-oncogene
The RET proto-oncogene encodes a tyrosine kinase receptor [75, 76], and RET activation promotes downstream signaling, leading to cell proliferation, differentiation and survival. [75]. Depending on the length of the C-terminus of the RET protein, three splice variants of the RET mRNA can be distinguished, namely, RET9, RET43 and RET51, and all present different cellular localization and function [77]. In PTC, gene fusions are the most common, but RET gene mutations were also associated with tumorigenesis, specifically RET G691S (rs1799939), L769 L (rs1800861) and S904S (rs1800863) [78]. Khan et al., suggested that rare variants G691SA and S904S are more prevalent in PTC and might be associated with a predisposition to TC development, as opposed to the underrepresented L769 L variant. However, this study was conducted on blood samples of post-thyroidectomy patients, thus the sensitivity of the assay remains to be determined.
Gene fusions
RET/PTC gene fusions
The variants of RET rearrangements are characterized by the fusion of the kinase domain to the 5′ terminus of the donor gene, resulting in a change of the subcellular localization of the receptor to the cytosol and leading to constitutive activation of the MAPK signaling pathway [79]. Until now, 25 fusion variants were described, 19 of which are associated with PTC [33, 80–92]. The RET kinase domain and 5′ end of CCD6 gene (RET/PTC1) fusion [84] or the nuclear receptor co-activator 4 gene (NCOA4) (RET/PTC3) are most common [81]. Zou et al., reported a 14% rate of RET/PTC rearrangement and co-occurrence of BRAFV600E with RAS/PTC1 (n = 82) [93]. Rossi et al., tested fine-needle aspiration of PTC samples by real-time PCR and showed that in 7.3% of the 940 samples, either RET/PTC1 or RET/PTC3 was present [37]. Six of the patients had both RET rearrangement and BRAF mutation. RET rearrangement appears to be fairly common in children with PTC [67]. Out of 13 samples in the study, RET gene fusions were detected in 2 (15%) samples by fluorescence in situ hybridization (FISH) assay.
KAZN–C1ORF196
Le Pennec et al., identified 4 novel gene fusions, most prominently KAZN–C1ORF196 [94], and this finding was confirmed in both a case study and in 85% of additional PTC samples (n = 94). KAZN encodes a keratinization-associated adhesion protein, whereas C1ORF196 is a putative gene. The biological function of such gene fusion is unknown, but it is predicted to be a result of an alternative splicing event generates a transcript coding for an in-frame protein. RNA sequencing of 115 samples from thyroid tumor tissues and metastases was performed, and 87 samples classified as PTC were sequenced using the Sanger method to validate the existing mutations [94]. KAZN–C1ORF196 gene fusion was absent in both tumor-adjacent (n = 37) and normal thyroid tissue (n = 23). Other mutations specific for the patients were identified, all of which highlight the tumor genetic heterogeneity. What is remarkable about this study is the fact that most of the mutations found were specific for a particular patient only.
Mutations of DNA-repair genes
CHEK2
Mutations in DNA repair genes appear to be mutually exclusive with MAPK activator mutations such as BRAFV600E, but they might exist simultaneously with other mutations involved in the MAPK signaling pathway, e.g., RAS (see below) [95]. Disruption of DNA repair can be a prognostic marker for aggressive PTC development, according to TCGA (See Table 2) [33]. Genotyping of a Polish population showed that 15.6% of samples (n = 468) had one of four cell cycle checkpoint kinase 2 (CHEK2) mutations known to contribute to carcinogenesis (truncating mutations IVS2 + 1G > A, 1100delC or del5395 and a mis-sense mutation I157T) [11]. Wójcicka et al., identified the rs17879961 variant as a risk allele for PTC in a group of 1781 patients (OR = 2.2, P = 2.37·10− 10) [96]. In a Greater Poland female population (case/control: 602/829), the c.470C (I157T) homozygous variant was shown to increase the risk of developing PTC by nearly 13-fold (OR = 12.81, P = 1.9·10− 2) and was observed in 3 women (0.57%), as determined by pyrosequencing [97]. A heterozygous variant of the same mutation increases the risk by 2-fold (OR = 1.7, P = 1.7·10− 2).This association was not observed for male patients.
Alterations in cell signaling pathways
RAS
Mutations in the family of RAS proteins are associated with AKT phosphorylation and result in preferential activation of the PI3K/AKT pathway in TC by evasion of apoptosis, proliferation and cellular growth [98, 99]. The RAS family consists of 4 proto-oncogenes: H-RAS, N-RAS, K-RAS4A and K-RAS4B [100]. Although RAS mutations are more prevalent in FTC, they are also observed in a subset of PTCs [101]. Zou et al., detected KRAS mutations (p.Q61R and p.S65 N) in 2 samples (2%, 2/88) and an NRAS (p.Q61R) mutation in 3 cases (PTC 1%, TCV 2%). Rossi et al., observed 3.4% of samples harboring a somatic RAS mutation (n = 940), which correlated with an aggressive histotype and poorer prognosis [37]. Until now, RAS mutations have not been found in juvenile thyroid tumors [67].
MUC1
Mucin (MUC1) plays a role in the signaling pathways of proliferation and differentiation of epithelial cells and is crucial in metastasis and tumorigenesis of epithelial cancers such as adenocarcinomas and ovarian cancer [102]. In PTC, MUC1 is thought to be a marker of poorer outcome (See Table 2), although this stance is controversial. Using pyrosequencing, Renaud et al., showed that 40% of 94 PTC samples overexpressed MUC1 in the cytoplasm, which correlated with the presence of the BRAFV600E mutation in 95% of samples.
Deregulation of protease expression
TMPRSS4
Transmembrane protease serine 4 (TMPRSS4) is a type II transmembrane serine protease overexpressed in several cancer types, including gastric [103], breast [104], lung [105] and thyroid cancers [105–107]. TMPRSS4 promotes cell proliferation, invasion, metastasis and epithelial-mesenchymal transition (EMT) and is predominantly overexpressed in PTC. Kebebew et al., tested 131 tumors by cDNA microarrays, and TMPRSS4 was one of the 6 genes deregulated in malignant tumors [107]. Jarząb et al., tested 50 samples from 33 patients (23 PTC, 10 other thyroid malignancies) paired with normal tissue using microarray analysis [106]. TMPRSS4 was classified as one of the genes forming a set of markers that distinguish between benign and malignant tumors.
Mutations in transcription regulators
EIF1AX
Eukaryotic translation initiation factor 1A/X-linked (EIF1AX) is a major player in the transfer of Met-tRNAf and has a high mutation rate in PTC (1.5%, 6/402). EIF1AX is suggested as a potential driver of tumorigenesis in other cancers, e.g., uveal melanoma [33, 108, 109], and in TC, it is a promising biomarker candidate. This observation is supported by Karanamurthy et al., who detected EIF1AX mutation in 2.3% (n = 3/86) of tested PTC samples and 1 of 5 PTC FNA samples using NGS [110]. Almost all of the EIF1AX mutations were located at a hotspot A113_splice site at intron 5/exon 6.
FOXE1
The thyroid transcription factor forkhead box E1 (FOXE1) possesses a well-conserved DNA binding domain (FDH) and is crucial in the development of a healthy thyroid [111]. Deregulation of transcription factors from the FOX family is recognized as an important element of TC progression.
Penna-Martinez et al., used PCR to genotype 196 PTC samples (German population) for the presence of two known susceptibility SNPs in FOXE1 [17, 112]. The rs965513 phenotypes “AA” and “AG” were more common in DTC patients in contrast to the “GG” phenotype, which was common in healthy controls. The rs965513 variant is more pronounced in PTC than in FTC [112]. Mond et al., sequenced 120 PTC tumors for SNPs in the coding region of FOXE1. Four mis-sense mutations were found in the FHD (c.821C > A, p.P54Q; c.943A > C p.K95Q; c.994C > T, p.L112F), each in a single tumor. Molecular modeling of the described mutations showed their location in a region highly conserved across species, thus explaining the potential carcinogenic effect [111].
TERT promoter
Telomerase reverse transcriptase (TERT) is a catalytic subunit of telomerase vital for the gain of immortality by cancer cells [113, 114]. Two mutations located in the TERT promoter region are associated with carcinogenesis, namely, C-to-T substitution (C1,295,228 T) and C-to-A substitution (C1,295,250A) [115]. TERT promoter mutations appear to be rare in PTC (4.4%, n = 455, Chinese population) [64], but they correlate positively with aggressiveness of the tumor and patient age (See Table 2). These results confirm studies performed by Liu et al., [116, 117]. TERT mutations are less common in PTC (11.3%, n = 408) than in ATC (42.6%, n = 54) when pooled data are considered [118]. Studies also show that TERT promoter mutations correlate with poorer outcomes and an increase in aggressiveness of the tumor, even if they do not coincide with BRAF mutation [115, 119]. TERT promoter mutations are most common in TCV.
Regulatory RNAs
RNA-mediated regulatory pathways disrupted in carcinogenesis involve micro-RNA (miRNA, miR) signaling. Micro-RNAs are short, 21–23 nt, non-coding endogenous RNA fragments that regulate expression at the posttranscriptional level [120]. MicroRNA-deregulated thyroid cancers are collected in Table 3. T Yoruker et al., used RT-PCR to test serum from pre- and post-operative PTC patients to measure the level of micro-RNA expression [121]. The PTC patient sera levels of 4 miRNAs (miR-222, miR-31, miR-151-5p, let-7) were significantly higher compared with healthy controls, and the miR-21 level was lower (see Table 2). General levels of all miRNAs were lower in the post-operative samples and showed no significant difference with the healthy control group. A similar study was performed by Lee et al., to measure the expression of miR-222 and miR-146b in plasma and tumor tissues [122]. In recurrent tumors, miRNAs were significantly up-regulated compared with non-recurrent patients and healthy controls. Plasma miRNAs levels decreased after thyroidectomy in both cases. The results, especially miR-222 overexpression, confirm the results of other groups [123, 124], suggesting that both miRNAs might be used as biomarkers of cancer progression. MiR-221, miR-22, and miR-21 are involved in PTEN regulation [125], whereas miR-126 is associated with angiogenesis [120], and its expression in PTCs as well as undifferentiated thyroid cancers showed a correlation between miR-126 down-regulation and overexpression of VEGF-A mRNA and protein in tumors. miR-639 expression was upregulated in cancer tissues [126]. In contrast, expression of miR-20b a regulator of the MAPK/ERK signaling pathway with potential tumor suppressor qualities, was down-regulated in TC [127]. Samsonov et al., showed the potential differentiating miRNAs (miR-21 and miR-181a) that might be useful in distinguishing PTC from FTC [128]. Studies conducted by Hu et al., associated down-regulation of miR-940, miR-15a, and miR-16 with PTC phenotype [129].
Table 3.
microRNAs differentially expressed in PTC and their tissue of origin
| Up-regulation | Localization | Sample origin | Down-regulation | Localization | Sample origin |
|---|---|---|---|---|---|
| let-7 | 19q13.41 | serum | miR-15a | 13q14.2 | tumor tissue |
| miR-31 | 9p21.3 | miR-16 | 13q14.2, 3q25.33 | ||
| miR-151-5p | 8q24.3 | miR-21 | 17q23.1 | serum | |
| miR-146b | 10q24.32 | plasma, tumor tissue | miR-126 | 9q34.3 | tumor tissue |
| miR-221 | Xp11.3 | miR-940 | 16p13.3 | ||
| miR-222 | Xp11.3 | ||||
| miR-639 | 19p13.12 | tumor tissue |
Follicular thyroid carcinoma (FTC)
Follicular thyroid carcinoma is the second most common thyroid malignancy, is considered more aggressive than PTC, and has a 95% 5-year survival rate. Mortality rate and disease aggressiveness increase with the age of the patient at diagnosis [130].
Hou et al., showed the occurrence of PTEN (7%, 6/86 samples) and PIK3CA (6%, 5/85 samples) mutations in FTC [42]. PIK3CA gene copy gain was found in 20% of tested samples (24/85). These mutations might affect the activation and regulation of the PI3K/Akt pathway. In contrast to PTC, the BRAFV600E mutation is generally rare in FTC [115]. TERT promoter mutations (see Table 4) were also tested, but the FTC sample number was low (20 minimally invasive FTCs without metastasis and 3 FTCs with metastasis). Nevertheless, the results correlated positively with the presence of distant metastases (1/2 minimally invasive samples with distant metastases).
Table 4.
Somatic mutations found in FTCs. SNV: Single nucleotide variant
| Gene | Localization | Gene function | Mutation | Defect in cancer | Clinial correlation | Literature |
|---|---|---|---|---|---|---|
| Gene | ||||||
| ARNT | 1q21.3 | N/A | CNV | unknown | N/A | Świerniak et al., 2016 |
| CHEK2 | 22q12.1 | protein kinase | SNV, (C29,108,001A) | gain of immortality | N/A | Świerniak et al., 2016, Wójcicka et al., 2014 |
| COL1A1 | 17q21.33 | pro-alpha1 chain of type I collagen | indel, chr17: 48275120 | unknown | N/A | Świerniak et al., 2016 |
| COX6/DERL2 | COX6/A1: 12q24.31, COX6/A2: 16p11.2, DERL2: 17p13.2 | N/A | translocation | unknown | N/A | Świerniak et al., 2016 |
| FBXW7 | 4q31.3 | subunit of ubiquitin protein ligase complex called SCFs | CNV | unknown | N/A | Świerniak et al., 2016 |
| FOXO4 | Xq13.1 | suppressor of transcription | SNV, (C70,321,204 T) | Deregulation of transcription, alters protein structure | N/A | Świerniak et al., 2016 |
| IDH1 | 2q34 | catalyzes the oxidative decarboxylation of isocitrate to 2-oxoglutarate | LOH | unknown | N/A | Świerniak et al., 2016 |
| JAK3 | 19p13.11 | Protein kinase | intronic region | unknown | N/A | Świerniak et al., 2016 |
| KAZN-C1ORF196 | KAZN: 1p36.21, C1ORF196: 1p36.21 | unknown | Gene fusion | unknown | N/A | Salajegheh et al., 2016 |
| KTN1 | 14q22.3 | membrane protein involved in organelle motility | deletion in chr14:56139994 | unknown | N/A | Świerniak et al., 2016 |
| MITF | 3p13 | transcription regulator | insertion, chr3:69987750 | unknown | N/A | Świerniak et al., 2016 |
| NCOA2 | 8q13.3 | epigenetic modifier | chr8 position 71,053,835 A > C | unknown | N/A | Świerniak et al., 2016 |
| PAX8/PPARG | PAX8: 2q14.1, PPARG: 3p25.2 | N/A | t(2;3)(q13;p25) translocation | competitor inhibitor of PPARγ/ transcription factor similar to endogenous PPARγ | N/A | Lacroix et al., 2005, Giordano et al., 2006 |
| PIK3CA | 3q26.32 | catalytic subunit of phosphatidylinositol 3-kinase | CNV (gain) | unknown | N/A | Hou et al., 2007 |
| TMPRSS4 | 11q23.3 | serine protease | overexpression | Promotes cancer cells proliferation, invasion and metastasis | positive correlation with staging of tumor nodes metastasis | Guan et al.,2015 |
| TERT promoter | 5p15.33 | telomerase reverse transcriptase | C228T (rs35809415), C250A, C250T(rs1020948523) | unknown | presence of metastases | Bae et al., 2016 |
| USP6 | 17p13.2 | ubiquitin Specific Peptidase | CNV | unknown | N/A | Świerniak et al., 2016 |
| WRN | 8p12 | repair od double stranded breaks | LOH | unknown | N/A | Świerniak et al., 2016 |
| Regulation of expression | ||||||
| miR-199a-5p | 19p13.2 | regulator of CTFG in healthy cells | Micro RNA | Disruption of regulatory pathways, propagation of cancer | Downregulation during tumorigenesis | Sun et al., 2016 |
Świerniak et al., performed targeted NGS sequencing of 48 FTC tumors [12]. The authors identified previously undescribed somatic mutations in both intronic and exonic regions. FTC mutations were found in FOXO4 (transcription suppressor), CHEK2 and NCOA2 (epigenetic modifier) genes. Additionally, 10/18 identified single nucleotide variants (SNVs) were located in the non-coding regions of the studied genes. Other types of mutations included indels in MITF and KTN1 genes (transcription factor and transmembrane kinesin receptor, respectively) and loss of heterozygosity (LOH) in the IDH1 gene that belongs to the dehydrogenase family. Copy number variations (CNV) in ARNT (facilitates transport to the nucleus, transcriptional co-regulator of HIF1 expression), FBXW7 (component of the ubiquitin degradation signaling chain) and USP6 (ubiquitin specific peptidase) were also found in samples with populations of cells highly represented in tumors. In the low-confidence FTC group, a distinct subset of mutations was found, meaning that the differentiation of the two subsets based on their molecular profiles might be possible. In lower-confidence FTC, subset mutations were found in the COL1A1 gene, which is a fibrin-forming type of collagen. LOHs were identified in WRN (belonging to a family of DNA and RNA helicases) and PPARγ (member of a nuclear receptor subfamily), among others. A new translocation of unknown function was described, namely, COX6C/DERL2. KAZN/C1ORF196 gene fusion was confirmed in the case study and in 55% (out of 11) of FTC additional samples [94].
One of the most common genetic events in follicular thyroid cancer is the gene fusion of PAX8/PPARγ or PPFP oncoprotein gene [131, 132]. PAX8 on its own is necessary for the normal development of the thyroid [133], and PPARγ is a nuclear receptor [134]. PAX8/PPARγ fusion is present in 35% of FTC tumors on average, can be overexpressed by up to 50-fold compared with endogenous PPARγ in tumor tissues [135, 136] and is probably the effector component of the oncogenic rearrangement [137].
In FTC, as in PTC, overexpression of TMPRSS4 is observed in 53.6% (15/28) of the samples, as shown by Guan et al. [138].
Sun et al., found a positive correlation between FTC tumorigenesis and low levels of miR-199a-5p expression [131]. MiR-199a-5p was identified as a regulator of the connective tissue growth factor (CTFG), which acts as an inhibitor of the cell cycle in healthy tissue. In tumor conditions, both fusion proteins appear to possess binding domains that retain their function in the correct cellular context [132].
Anaplastic thyroid carcinoma (ATC)
Anaplastic thyroid carcinoma is the most aggressive type of TC and contributes to 1–2% of all thyroid cancers and 39% of reported deaths [133]. The 6- to 12-month mortality rates reach 80%. The high aggressiveness of ATC is caused by dedifferentiation of well-differentiated thyroid cancer forms such as PTC [134–136]. Compared with PTC and poorly differentiated thyroid cancers, the mutation burden in ATC is much larger [137] (see Table 5).
Table 5.
Somatic mutations found in ATCs
| Gene | Localization | Gene function | Mutation | Defect in cancer | Physiological effect | Literature |
|---|---|---|---|---|---|---|
| Chromosomal Abberation | ||||||
| KAZN-CIORF196 | 1p36.21, 1p36.21 | N/A | Gene fusion | potential role in progression and development of tumors | Le Pennec et al., 2015 | |
| Gene | ||||||
| ARID1A, ARID1B, ARD2, ARID5B, SMARCB1, PBRM1, ATRX | 1p36.11, 6q25.3, N/A, N/A, 22q11.23, 3p21.1, Xq21.1 | components of the SWI/SNF complex, responsible for the chromatin remodeling | N/A | mutation in one of the complex components leads to dysfunction of the whole complex | N/A | Landa et al., 2016, Latteyer et al., 2016 |
| ALK | 2p23.1 | anaplastic lymphoma kinase | D1203H | hallmark of anaplastic tumors | N/A | Bonhomme et al., 2017, Latteyer et al., 2016 |
| ATM | 11q22.3 | cell-cycle checkpoint, response to DNA damage | E2039K | higher mutation burden, consistent with the lack of checkpoint function | N/A | Landa et al.,2016, Kunstman et al.,2015 |
| BRAF rs113488022 | 7q34 | serine/threonine kinase, response to cell growth factors | V600E | constitutive activation of MAPK pathway | N/A | Santarpia et al., 2008, Guerra et al., 2013, Kasaian et al., 2015, Landa et al., 2016, Latteyer et al., 2016 |
| DAXX | 6p21.32 | transcription repressor binding the sumoylated transcription factors | S641X | potential driver mutation | correlates with non-thyroidal malignancies | Kunstman et al., 2015 |
| EIF1AX | Xp22.12 | translation initiation factor, transfer of met-trnaf | Splice site 1 bp upstream of ex6 (C > G), G9R (C > G), P2R(G > C) | potential driver mutation | N/A | Kunstman et al., 2015, Landa et al., 2016 |
| ERBB2 | 17q12 | downstream enhancer of kinase-mediated signaling pathways | D387N | potential driver mutation | N/A | Kunstman et al., 2015 |
| D873N, A763T | N/A | Bonhomme et al., 2017 | ||||
| HECTD1 rs769574276 | 14q12 | ubiquitin protein ligase | L547 V | impairment of ubiquitynylated proteins degradation | N/A | Kunstman et al., 2015 |
| KMT2A, KMT2C, KMT2D (MLL2), SETD2 | 11q23.3, 7q36.1, 12q13.12, 3p21.31 | histone methyltransferases, epigenetic modifiers | N/A, KMT2D: Q1892Q (rs753626919), R5389W | impairment of epigenetic mechanisms, potential driver mutation | N/A, KMT2D: correlates with non-thyroidal malignancies | Landa et al., 2016, Kunstman et al., 2015 |
| MET | 7q31.2 | tyrosine-protein kinase met | I166T | proto-oncogene | N/A | Bonhomme et al., 2017 |
| mTOR | 1p36.22 | response element = to stress, possessing kinase activity | R164Q (rs573705289), M2327I | potential driver mutation | correlates with non-thyroidal malignancies | Kunstman et al., 2015 |
| NF1 | 17q11.2 | neurofibromatosis related gene | P2696L (rs778799019), R2496X (rs752162999) | potential driver mutation | correlates with non-thyroidal malignancies | Kunstman et al., 2015, Landa et al., 2016, Latteyer et al., 2016 |
| NOTCH1–4 (NOTCH2 in Kunstman) | 1p12 | transmembrane receptors | NOTCH2: S361F (rs587735797), R1393H | potential driver mutation | correlates with non-thyroidal malignancies | Kunstman et al., 2015, Landa et al., 2016 |
| PIK3CA | 3q26.32 | PI3K/AKT/mTOR pathway effector | E542K (rs121913273), E545K (rs104886003) | mutation of helical domain | N/A | Landa et al., 2016, Kunstman et al., 2015, Hou et al., 2007 |
| PTEN | 10q23.31 | PI3K/AKT/mTOR pathway effector | N/A | truncated protein | N/A | Landa et al., 2016, Hou et al., 2007 |
| RAS | 11p15.5, 1p13.2, 12p12.1 | signal transduction | N/A | preferential activation of PI3K-AKT pathway | N/A | Santarpia et al., 2008, Guerra et al., 2013, Landa et al., 2016, Latteyer et al., 2016, Hou et al., 2007 |
| TERT promoter | 5p15.33 | telomerase reverse transcriptase | C228T (rs35809415), C250T (rs1020948523) | gain of immortality | shorter survival | Bae et al., 2016, Landa et al., 2016 |
| TMPRSS4 | 11q23.3 | serine protease | N/A | promotes proliferation | positive correlation with tumor grade | Guan et al., 2015 |
| TP53 | 17p13.1 | tumor suppressor protein | Y163C (rs148924904) | gain of immortality | N/A | Kasaian et al., 2015, Landa et al., 2016, Bonhomme et al., 2017 |
| USH2A | 1q41 | uscherin, extracellular matrix binding protein interacting with collagen and fibronectin | I2189V (rs542406401), D798V (rs148431156), E571K(C > T), L1727F(G > A) | missense mutations | N/A | Kunstman et al., 2015 |
| CREBBP | 16p13.3 | histone acetyltransferase | N/A | epigenetic modifier | N/A | Landa et al., 2016 |
| EP300, BCOR, BCL6 | 22q13.2, Xp11.4, 3q27.3 | epigenetic modifiers | N/A | abnormal protein modifications | N/A | Landa et al., 2016 |
| CTNNB1 | 3p22.1 | cytoskeletal anchor, adhesive junctions | Q108H | unknown | N/A | Kunstman et al., 2015, Landa et al., 2016 |
| MSH2 | 2p21 | DNA mismatch repair | N/A | gain of mutation phenotype | N/A | Landa et al., 2016, Kunstman et al., 2015 |
| MSH5 | 6p21.33 | A199V (C > T) | N/A | |||
| MSH6 | 2p16.3 | D736H (G > C) | N/A | |||
| MLH1 | 3p22.2 | I19M (C > G), I68M (rs780141938), Q60X (C > T) | N/A | |||
| MLH3 | 14q24.3 | L264 V (G > C) | N/A | |||
ATC can arise independently, but it often coincides with well-differentiated tumors. Co-occurrence of BRAF and RAS mutations in ATC suggests its common genetic origin with DTC [135, 139, 140]. Hou et al., tested 50 ATC tumors and found a high prevalence of mutations associated with PI3K/Akt pathway activation: PTEN 16% (8/50) and PIK3CA 12% (6/50) [42]. RAS mutations were also identified in 8% (4/50) of samples. The molecular heterogeneity of ATC makes it incredibly difficult to analyze. Kasaian et al., performed whole-genome sequencing of 1 ATC sample and identified 24 somatic mutations, including two heterozygous mutations in BRAF (V600E) and TP53 (Y163C) genes. [141]. Kunstman et al., tested 22 tumor samples with whole-exome sequencing [142]. The majority (68%) of the observed variants code for mis-sense mutations. A total of 16 genes were identified as potential drivers of tumorigenesis, 6 of which were present in multiple samples, namely, NF1 (negative regulator of RAS pathway), mTOR (kinase, mediates response to stress), ERBB2 (EGF receptor), DAXX (apoptosis regulator and transcription repressor among other functions), MLL2 (histone methyltransferase), and NOTCH2 (regulator of cell fate). In addition, recurrent mutations of EIF1AX and HECTD1 (ubiquitin-transferase activity) and non-synonymous USH2A (development of retina and inner ear) mutations were observed. Several of the tested cases presented a hypermutation phenotype, resulting in a high mutation burden of mismatch repair genes. Bonhomme et al., sequenced 94 ATC tumors targeted to TERT using NGS and 98 samples using Sanger sequencing [143]. More than 50% of samples possessed the TP53 mutations, and ALK rearrangements were rare. In total, 210 different alterations were found, including those not previously described in the context of TC, such as MET (proto-oncogene) and ERBB2 mutations. In the Korean population, 60% samples (3/5) had a TERT promoter mutation, which coincided with BRAFV600E [115]. In a study by Landa et al., the presence of BRAFV600E mutation was observed in 45% out of 33 tumors [137]. In the same study RAS mutations (H-RAS, K-RAS, and N-RAS) occurred in 24% of the samples but were mutually exclusive with BRAFV600E.
Other mutations found in ATCs were NF1 (3 samples), PIK3CA (18%), and PTEN (15%). PIK3CA mutation tends to co-occur with BRAF mutations, whereas NF1 tends to be present simultaneously with PTEN mutations. EIF1AX mutations were present in 9% of the 33 studied tumors.
For the first time, Landa et al. reported mutations in components of the SWI/SNF complex (chromatin remodeling system), as reported in 36% (n = 33) of tumors. Mutations were also found in histone methyltransferase genes (KMT2A, KMT2C, KMT2D, and SETD2) in 24% (n = 33) of ATCs. Additional genes involved in epigenetic processes, i.e., CREBBP, EP300, BCOR, and BCL6, were mutated at low frequencies. One sample carried a CTNNB1 (p.L347P; WNT signaling pathway) mutation, but this finding was not validated by others. Mutations were also observed in members of the MMR DNA repair pathway (MSH2, MSH6, and MLH1) in 12% of samples. Another DNA damage response element, ATM, was mutated in 9% of tested ATCs. Landa et al., reported frequent (73%, n = 33) TERT promoter and TP53 mutations. The TERT promoter C228T variant was more common than the C250T variant. TERT promoter mutations significantly diminished the survival rate from 732 to 147 days.
Gene fusions are also present in ATC. KAZN/C1ORF196 was identified by Le Pennec et al., in a case study and confirmed in 11% of additional ATC samples [94]. Guan et al., observed an increase of TMPRSS4 expression in all ATC samples (n = 12) compared with adjacent normal tissue [138]. Targeted DNA sequencing for TP53, RAS, BRAF, ALK, and NF1 of 30 formalin-fixed paraffin-embedded (FFPE) ATC tumor samples by Latteyer et al., showed that 28/30 tested samples carried at least one of the tested mutations [144]. TP53 mutation was most common (18/30), followed by NF1 (11/30) and RAS family mutations (7/30 combined). It is also worth mentioning that nearly a third of the samples showed residual contaminations of either PTC or FTC tissue, proving the anaplastic tumor heterogeneity.
Zhang et al., tested the expression of myocardin family genes (involved in cell growth arrest, inhibition of differentiation, metastasis and tumor invasion) [145]. MRTF-A was overexpressed in metastatic ATC but was not present in either in primary tumor or the adjacent tissue. Following this finding, down-regulation of miR-206 was identified as the factor leading to the MRTF-A overexpression.
Conclusions
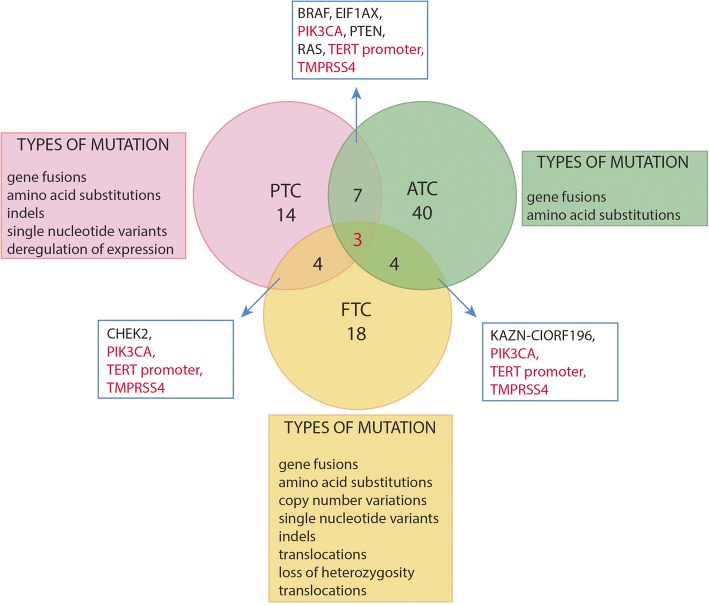
Despite the large number of mutations involved in the tumorigenesis of thyroid carcinomas (Fig. 2), many tumors remain unclassified by FNA biopsy or even genetic testing. Pagan et al., notes that over 50% of samples tested for a large number of reported mutations already observed in TC by RNA-seq do not show a phenotype, leading to the conclusion that the fast-growing database of somatic and driver mutations in thyroid cancers must be expanded with respect to histological subtype [146].
Fig. 2.
Genetic changes identified in thyroid cancers of follicular origin. Genes common for all three TC subtypes are marked in red
DNA methylation in thyroid cancer has been extensively studied and reviewed but was not discussed in detail in this review. However, it is worth mentioning that the advances in next-generation sequencing and microarray techniques enable in-depth research on the methylation pattern in GC-rich regions and its effect on gene expression. Most studies focus on pre-determined loci [147, 148], and fewer are available at the whole-genome scale [149, 150]. Determination of the methylation patterns can be potentially useful for differentiating between TC subtypes with greater precision. The largest study to date that examines whole-genome methylation was performed as a component of the TCGA project (PTC, n = 496) [33]. In a recent study, Bisarro dos Reis et al., proposed a hyper/hypomethylation genetic signature that allows distinction between TC subtypes (Hürtle cell, PTC, FTC, non-neoplastic tissue and benign lesions, ATC) based on the Illumina 45 k platform, with high sensitivity and specificity (63 and 92%, respectively) [151]. Methylation can also be used as a prognostic marker of disease outcome, as proposed in the same article. Beltrami et al., proposed the PTC hypomethylation signature of 41 PTC-paired samples (88% of hypomethylation) as a prognostic biomarker of PTC development [152]. This signature coincides with the presence of the BRAFV600E mutation (68% of the hypomethylation signature).
In the era of advanced molecular analysis, genetic markers have become a useful tool for the evaluation of thyroid tumor growth and progression. Molecular biomarkers can be applied in the classification of thyroid tumor subtypes and the prediction of disease outcome and might also aid development of systemic molecular therapies in cancers that are refractory to standard treatment. The discovery of specific genetic alterations and mechanisms of thyroid carcinoma development is expected to lead to more personalized treatment for patients with advanced and recurrent disease. Despite the presence of the molecular changes described in this review, the roles of molecular biomarkers in the development of different thyroid tumor subtypes still remain unclear.
Availability of data and materials
See section “References”.
Abbreviations
- ATC
Anaplastic thyroid cancer
- CNV
Copy number variations
- DTC
Differentiated thyroid cancer
- FISH
Fluorescence in situ hybridization
- FNAB
Fine-needle aspiration biopsy
- FTC
Follicular thyroid cancer
- GWAS
Genome-wide association study
- HRM
High resolution melting
- LOH
Loss of heterozygosity
- MTC
Medullary thyroid cancer
- NGS
Next-generation sequencing
- PCR
Polymerase chain reaction
- PTC
Papillary thyroid cancer
- SNP
Single nucleotide polymorphism
- SNV
Single nucleotide variant
- TC
Thyroid cancer
- TCGA
The Cancer Genome Atlas
- TCV
Tall cell variant
Authors’ contributions
NP – has contributed substantially to the concept of the manuscript, researched and analyzed the literature data, was a major contributor in writing the review article, has been involved in revising the manuscript critically for important intellectual content. KZ – has contributed to the concept of the manuscript, has been involved in revising the manuscript critically for important intellectual content. HARB – has contributed to the concept of the manuscript, has been involved in revising the manuscript critically for important intellectual content. JW – has contributed substantially to the concept of the manuscript, has been involved in drafting the manuscript and revising it critically for important intellectual content, contributed to writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
See section “References”, approvals within particular articles included in the literature search.
Consent for publication
not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Natalia Pstrąg, Email: natalia.pstrag@amu.edu.pl.
Katarzyna Ziemnicka, Email: kaziem@ump.edu.pl.
Hans Bluyssen, Email: h.bluyss@amu.edu.pl.
Joanna Wesoły, Email: j.wesoly@amu.edu.pl.
References
- 1.NationalCancerInstitute. Available from: https://seer.cancer.gov/statfacts/html/thyro.html.
- 2.Carneiro RM, et al. Targeted therapies in advanced differentiated thyroid cancer. Cancer Treat Rev. 2015;41(8):690–8. doi: 10.1016/j.ctrv.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Hoang JK, Nguyen XV, Davies L. Overdiagnosis of thyroid cancer: answers to five key questions. Acad Radiol. 2015;22(8):1024–1029. doi: 10.1016/j.acra.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 4.CancerResearchUK. Thyroid cancer survival statistics. Available from: http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/thyroid-cancer/survival#heading-Zero.
- 5.American Cancer Society, Cancer Facts & Figures. Antlanta: American Cancer Society, 2017. 2017.
- 6.Arribas J, et al. Expression of YY1 in differentiated thyroid Cancer. Endocr Pathol. 2015;26(2):111–118. doi: 10.1007/s12022-015-9359-6. [DOI] [PubMed] [Google Scholar]
- 7.Nagy R, Ringel MD. Genetic predisposition for nonmedullary thyroid cancer. Horm Cancer. 2015;6(1):13–20. doi: 10.1007/s12672-014-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiacchio S, et al. Anaplastic thyroid cancer: prevalence, diagnosis and treatment. Minerva Endocrinol. 2008;33(4):341–357. [PubMed] [Google Scholar]
- 9.Gennaro Chiappetta TV, Vitiello M, Pasquinelli R, Monaco M, Palma G, Sepe R, Luciano A, Pallante P, Palmieri D, Aiello C, Rea D, Losito SN, Arr C, Fusco A, Fedele M. PATZ1 acts as a tumor supressor in thyroid cancer via targeting p53-dependent genes involved in EMT and cell migration. Oncotarget. 2014;6(7):14. doi: 10.18632/oncotarget.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodish MB, Stratakis CA. RET oncogene in MEN2, MEN2B, MTC and other forms of thyroid cancer. Expert Rev Anticancer Ther. 2008;8(4):625–632. doi: 10.1586/14737140.8.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siolek M, et al. CHEK2 mutations and the risk of papillary thyroid cancer. Int J Cancer. 2015;137(3):548–552. doi: 10.1002/ijc.29426. [DOI] [PubMed] [Google Scholar]
- 12.Swierniak M, et al. Somatic mutation profiling of follicular thyroid cancer by next generation sequencing. Mol Cell Endocrinol. 2016;433:130–137. doi: 10.1016/j.mce.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Rebhan M, et al. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13(4):163. doi: 10.1016/S0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 14.Figlioli G, et al. Novel genome-wide association study-based candidate loci for differentiated thyroid cancer risk. J Clin Endocrinol Metab. 2014;99(10):E2084–E2092. doi: 10.1210/jc.2014-1734. [DOI] [PubMed] [Google Scholar]
- 15.Kohler A, et al. Genome-wide association study on differentiated thyroid cancer. J Clin Endocrinol Metab. 2013;98(10):E1674–E1681. doi: 10.1210/jc.2013-1941. [DOI] [PubMed] [Google Scholar]
- 16.Gudmundsson J, et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44(3):319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudmundsson J, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41(4):460–4. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereda CM, et al. Common variants at the 9q22.33, 14q13.3 and ATM loci, and risk of differentiated thyroid cancer in the Cuban population. BMC Genet. 2015;16:22. doi: 10.1186/s12863-015-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. Association between XRCC1 and XRCC3 gene polymorphisms and risk of thyroid cancer. Int J Clin Exp Pathol. 2015;8(3):3160–3167. [PMC free article] [PubMed] [Google Scholar]
- 20.Papadakis M, et al. Follicular variant of papillary thyroid cancer in Alstrom syndrome. Familial Cancer. 2015;14(4):599–602. doi: 10.1007/s10689-015-9816-x. [DOI] [PubMed] [Google Scholar]
- 21.Bastos HN, et al. Association of polymorphisms in genes of the homologous recombination DNA repair pathway and thyroid cancer risk. Thyroid. 2009;19(10):1067–1075. doi: 10.1089/thy.2009.0099. [DOI] [PubMed] [Google Scholar]
- 22.Fayaz S, et al. Increased risk of differentiated thyroid carcinoma with combined effects of homologous recombination repair gene polymorphisms in an Iranian population. Asian Pac J Cancer Prev. 2014;14(11):6727–6731. doi: 10.7314/APJCP.2013.14.11.6727. [DOI] [PubMed] [Google Scholar]
- 23.Jones AM, et al. Thyroid cancer susceptibility polymorphisms: confirmation of loci on chromosomes 9q22 and 14q13, validation of a recessive 8q24 locus and failure to replicate a locus on 5q24. J Med Genet. 2012;49(3):158–163. doi: 10.1136/jmedgenet-2011-100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damiola F, et al. Contribution of ATM and FOXE1 (TTF2) to risk of papillary thyroid carcinoma in Belarusian children exposed to radiation. Int J Cancer. 2014;134(7):1659–1668. doi: 10.1002/ijc.28483. [DOI] [PubMed] [Google Scholar]
- 25.Liyanarachchi S, et al. Cumulative risk impact of five genetic variants associated with papillary thyroid carcinoma. Thyroid. 2013;23(12):1532–1540. doi: 10.1089/thy.2013.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullock M, et al. Association of FOXE1 polyalanine repeat region with papillary thyroid cancer. J Clin Endocrinol Metab. 2012;97(9):E1814–9. doi: 10.1210/jc.2012-1456. [DOI] [PubMed] [Google Scholar]
- 27.Maillard, S., et al., Common variants at 9q22.33, 14q13.3, and ATM loci, and risk of differentiated thyroid cancer in the French Polynesian population. PLoS One, 2015. 10(4): p. e0123700. [DOI] [PMC free article] [PubMed]
- 28.Nikiforov YE, et al. Highly accurate diagnosis of cancer in thyroid nodules with follicular neoplasm/suspicious for a follicular neoplasm cytology by ThyroSeq v2 next-generation sequencing assay. Cancer. 2014;120(23):3627–3634. doi: 10.1002/cncr.29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikiforov YE, et al. Impact of the multi-gene ThyroSeq next-generation sequencing assay on Cancer diagnosis in thyroid nodules with atypia of undetermined significance/follicular lesion of undetermined significance cytology. Thyroid. 2015;25(11):1217–1223. doi: 10.1089/thy.2015.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikiforova MN, et al. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98(11):E1852–E1860. doi: 10.1210/jc.2013-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudmundsson J, et al. A genome-wide association study yields five novel thyroid cancer risk loci. Nat Commun. 2017;8:14517. doi: 10.1038/ncomms14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cipollini M, et al. Polymorphisms within base and nucleotide excision repair pathways and risk of differentiated thyroid carcinoma. DNA Repair (Amst) 2016;41:27–31. doi: 10.1016/j.dnarep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research, N., Integrated genomic characterization of papillary thyroid carcinoma. Cell, 2014. 159(3): p. 676–90. [DOI] [PMC free article] [PubMed]
- 34.Kimura ET, et al. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63(7):1454–7. [PubMed] [Google Scholar]
- 35.Melillo RM, et al. The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells. J Clin Invest. 2005;115(4):1068–1081. doi: 10.1172/JCI200522758. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Xing M, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab. 2005;90(12):6373–6379. doi: 10.1210/jc.2005-0987. [DOI] [PubMed] [Google Scholar]
- 37.Rossi M, et al. Relevance of BRAF(V600E) mutation testing versus RAS point mutations and RET/PTC rearrangements evaluation in the diagnosis of thyroid cancer. Thyroid. 2015;25(2):221–228. doi: 10.1089/thy.2014.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson YC, et al. High rate of BRAF and RET/PTC dual mutations associated with recurrent papillary thyroid carcinoma. Clin Cancer Res. 2009;15(2):485–491. doi: 10.1158/1078-0432.CCR-08-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Cristofaro J, et al. Molecular genetic study comparing follicular variant versus classic papillary thyroid carcinomas: association of N-ras mutation in codon 61 with follicular variant. Hum Pathol. 2006;37(7):824–830. doi: 10.1016/j.humpath.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Z, et al. Prevalence of RET/PTC rearrangements in thyroid papillary carcinomas: effects of the detection methods and genetic heterogeneity. J Clin Endocrinol Metab. 2006;91(9):3603–3610. doi: 10.1210/jc.2006-1006. [DOI] [PubMed] [Google Scholar]
- 41.Costa AM, et al. BRAF mutation associated with other genetic events identifies a subset of aggressive papillary thyroid carcinoma. Clin Endocrinol. 2008;68(4):618–634. doi: 10.1111/j.1365-2265.2007.03077.x. [DOI] [PubMed] [Google Scholar]
- 42.Hou P, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13(4):1161–1170. doi: 10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- 43.Shattuck TM, et al. Independent clonal origins of distinct tumor foci in multifocal papillary thyroid carcinoma. N Engl J Med. 2005;352(23):2406–2412. doi: 10.1056/NEJMoa044190. [DOI] [PubMed] [Google Scholar]
- 44.Kaliszewski K, et al. Multi- and Unifocal thyroid microcarcinoma: are there any differences? Adv Clin Exp Med. 2016;25(3):485–492. doi: 10.17219/acem/62327. [DOI] [PubMed] [Google Scholar]
- 45.Barczynski M, et al. Prophylactic central neck dissection for papillary thyroid cancer. Br J Surg. 2013;100(3):410–418. doi: 10.1002/bjs.8985. [DOI] [PubMed] [Google Scholar]
- 46.Kim HJ, et al. Multifocality, but not bilaterality, is a predictor of disease recurrence/persistence of papillary thyroid carcinoma. World J Surg. 2013;37(2):376–384. doi: 10.1007/s00268-012-1835-2. [DOI] [PubMed] [Google Scholar]
- 47.Kiriakopoulos A, Petralias A, Linos D. Multifocal versus solitary papillary thyroid carcinoma. World J Surg. 2016;40(9):2139–2143. doi: 10.1007/s00268-016-3628-5. [DOI] [PubMed] [Google Scholar]
- 48.Giannini R, et al. The heterogeneous distribution of BRAF mutation supports the independent clonal origin of distinct tumor foci in multifocal papillary thyroid carcinoma. J Clin Endocrinol Metab. 2007;92(9):3511–3516. doi: 10.1210/jc.2007-0594. [DOI] [PubMed] [Google Scholar]
- 49.Kuhn E, et al. Different clonal origin of bilateral papillary thyroid carcinoma, with a review of the literature. Endocr Pathol. 2012;23(2):101–107. doi: 10.1007/s12022-012-9202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugg SL, et al. Distinct multiple RET/PTC gene rearrangements in multifocal papillary thyroid neoplasia. J Clin Endocrinol Metab. 1998;83(11):4116–4122. doi: 10.1210/jcem.83.11.5271. [DOI] [PubMed] [Google Scholar]
- 51.de Biase D, et al. High-sensitivity BRAF mutation analysis: BRAF V600E is acquired early during tumor development but is heterogeneously distributed in a subset of papillary thyroid carcinomas. J Clin Endocrinol Metab. 2014;99(8):E1530–8. doi: 10.1210/jc.2013-4389. [DOI] [PubMed] [Google Scholar]
- 52.Park SY, et al. Analysis of differential BRAF(V600E) mutational status in multifocal papillary thyroid carcinoma: evidence of independent clonal origin in distinct tumor foci. Cancer. 2006;107(8):1831–8. doi: 10.1002/cncr.22218. [DOI] [PubMed] [Google Scholar]
- 53.Jovanovic L, et al. Most multifocal papillary thyroid carcinomas acquire genetic and morphotype diversity through subclonal evolution following the intra-glandular spread of the initial neoplastic clone. J Pathol. 2008;215(2):145–154. doi: 10.1002/path.2342. [DOI] [PubMed] [Google Scholar]
- 54.Lin X, et al. Molecular analysis of multifocal papillary thyroid carcinoma. J Mol Endocrinol. 2008;41(4):195–203. doi: 10.1677/JME-08-0063. [DOI] [PubMed] [Google Scholar]
- 55.McCarthy RP, et al. Molecular evidence for the same clonal origin of multifocal papillary thyroid carcinomas. Clin Cancer Res. 2006;12(8):2414–8. doi: 10.1158/1078-0432.CCR-05-2818. [DOI] [PubMed] [Google Scholar]
- 56.Moniz S, et al. Clonal origin of non-medullary thyroid tumours assessed by non-random X-chromosome inactivation. Eur J Endocrinol. 2002;146(1):27–33. doi: 10.1530/eje.0.1460027. [DOI] [PubMed] [Google Scholar]
- 57.Wang W, et al. Clonal analysis of bilateral, recurrent, and metastatic papillary thyroid carcinomas. Hum Pathol. 2010;41(9):1299–1309. doi: 10.1016/j.humpath.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Abdel-Mageed AB, Kandil E. BRAF mutation in papillary thyroid carcinoma. Int J Clin Exp Med. 2012;5(4):310–315. [PMC free article] [PubMed] [Google Scholar]
- 59.Ozgursoy OB, Eisele DW, Tufano RP. The prognostic implications from molecular testing of thyroid cancer. Otolaryngol Clin N Am. 2014;47(4):595–607. doi: 10.1016/j.otc.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 60.Walts AE, et al. BRAF genetic heterogeneity in papillary thyroid carcinoma and its metastasis. Hum Pathol. 2014;45(5):935–941. doi: 10.1016/j.humpath.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Fnais N, et al. Diagnostic value of fine needle aspiration BRAF(V600E) mutation analysis in papillary thyroid cancer: a systematic review and meta-analysis. Hum Pathol. 2015;46(10):1443–1454. doi: 10.1016/j.humpath.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao T, et al. BRAF-activated LncRNA functions as a tumor suppressor in papillary thyroid cancer. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoo SK, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet. 2016;12(8):e1006239. doi: 10.1371/journal.pgen.1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kimbrell HZ, et al. BRAF testing in multifocal papillary thyroid carcinoma. Biomed Res Int. 2015;2015:486391. doi: 10.1155/2015/486391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun J, et al. BRAF V600E and TERT promoter mutations in papillary thyroid carcinoma in Chinese patients. PLoS One. 2016;11(4):e0153319. doi: 10.1371/journal.pone.0153319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gertz RJ, et al. Mutation in BRAF and other members of the MAPK pathway in papillary thyroid carcinoma in the pediatric population. Arch Pathol Lab Med. 2016;140(2):134–139. doi: 10.5858/arpa.2014-0612-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Z, et al. Clonality analysis of multifocal papillary thyroid carcinoma by using genetic profiles. J Pathol. 2016;239(1):72–83. doi: 10.1002/path.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gandolfi G, et al. Allele percentage of the BRAF V600E mutation in papillary thyroid carcinomas and corresponding lymph node metastases: no evidence for a role in tumor progression. J Clin Endocrinol Metab. 2013;98(5):E934–E942. doi: 10.1210/jc.2012-3930. [DOI] [PubMed] [Google Scholar]
- 70.Ming J, et al. Association between BRAF and RAS mutations, and RET rearrangements and the clinical features of papillary thyroid cancer. Int J Clin Exp Pathol. 2015;8(11):15155–15162. [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J, et al. Lymphatic mapping establishes the role of BRAF gene mutation in papillary thyroid carcinoma. Ann Surg. 2006;244(5):799–804. doi: 10.1097/01.sla.0000224751.80858.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guerra A, et al. The primary occurrence of BRAF(V600E) is a rare clonal event in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012;97(2):517–524. doi: 10.1210/jc.2011-0618. [DOI] [PubMed] [Google Scholar]
- 73.Lee MY, et al. Genetic alterations and their clinical implications in high-recurrence risk papillary thyroid Cancer. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, et al. High prevalence and mutual exclusivity of genetic alterations in the phosphatidylinositol-3-kinase/akt pathway in thyroid tumors. J Clin Endocrinol Metab. 2007;92(6):2387–2390. doi: 10.1210/jc.2006-2019. [DOI] [PubMed] [Google Scholar]
- 75.Arighi E, Borrello MG, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 2005;16(4–5):441–467. doi: 10.1016/j.cytogfr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 76.Ibanez, C.F., Structure and physiology of the RET receptor tyrosine kinase. Cold Spring Harb Perspect Biol, 2013. 5(2). [DOI] [PMC free article] [PubMed]
- 77.Myers SM, et al. Characterization of RET proto-oncogene 3′ splicing variants and polyadenylation sites: a novel C-terminus for RET. Oncogene. 1995;11(10):2039–2045. [PubMed] [Google Scholar]
- 78.Khan MS, et al. Possible impact of RET polymorphism and its Haplotypic association modulates the susceptibility to thyroid Cancer. J Cell Biochem. 2015;116(8):1712–8. doi: 10.1002/jcb.25130. [DOI] [PubMed] [Google Scholar]
- 79.Romei C, Ciampi R, Elisei R. A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat Rev Endocrinol. 2016;12(4):192–202. doi: 10.1038/nrendo.2016.11. [DOI] [PubMed] [Google Scholar]
- 80.Grieco M, et al. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60(4):557–563. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- 81.Santoro M, et al. Molecular characterization of RET/PTC3; a novel rearranged version of the RETproto-oncogene in a human thyroid papillary carcinoma. Oncogene. 1994;9(2):509–516. [PubMed] [Google Scholar]
- 82.Bongarzone I, et al. Molecular characterization of a thyroid tumor-specific transforming sequence formed by the fusion of ret tyrosine kinase and the regulatory subunit RI alpha of cyclic AMP-dependent protein kinase a. Mol Cell Biol. 1993;13(1):358–366. doi: 10.1128/MCB.13.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciampi R, Nikiforov YE. RET/PTC rearrangements and BRAF mutations in thyroid tumorigenesis. Endocrinology. 2007;148(3):936–941. doi: 10.1210/en.2006-0921. [DOI] [PubMed] [Google Scholar]
- 84.Corvi R, et al. RET/PCM-1: a novel fusion gene in papillary thyroid carcinoma. Oncogene. 2000;19(37):4236–4242. doi: 10.1038/sj.onc.1203772. [DOI] [PubMed] [Google Scholar]
- 85.Fugazzola L, et al. Molecular and biochemical analysis of RET/PTC4, a novel oncogenic rearrangement between RET and ELE1 genes, in a post-Chernobyl papillary thyroid cancer. Oncogene. 1996;13(5):1093–1097. [PubMed] [Google Scholar]
- 86.Hamatani K, et al. A novel RET rearrangement (ACBD5/RET) by pericentric inversion, inv(10)(p12.1;q11.2), in papillary thyroid cancer from an atomic bomb survivor exposed to high-dose radiation. Oncol Rep. 2014;32(5):1809–1814. doi: 10.3892/or.2014.3449. [DOI] [PubMed] [Google Scholar]
- 87.Klugbauer S, et al. Detection of a novel type of RET rearrangement (PTC5) in thyroid carcinomas after Chernobyl and analysis of the involved RET-fused gene RFG5. Cancer Res. 1998;58(2):198–203. [PubMed] [Google Scholar]
- 88.Klugbauer S, et al. A novel type of RET rearrangement (PTC8) in childhood papillary thyroid carcinomas and characterization of the involved gene (RFG8) Cancer Res. 2000;60(24):7028–7032. [PubMed] [Google Scholar]
- 89.Klugbauer S, Rabes HM. The transcription coactivator HTIF1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene. 1999;18(30):4388–4393. doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- 90.Nakata T, et al. Fusion of a novel gene, ELKS, to RET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosomes Cancer. 1999;25(2):97–103. doi: 10.1002/(SICI)1098-2264(199906)25:2<97::AID-GCC4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 91.Saenko V, et al. Novel tumorigenic rearrangement, Delta rfp/ret, in a papillary thyroid carcinoma from externally irradiated patient. Mutat Res. 2003;527(1–2):81–90. doi: 10.1016/S0027-5107(03)00056-3. [DOI] [PubMed] [Google Scholar]
- 92.Salassidis K, et al. Translocation t(10;14)(q11.2:q22.1) fusing the kinetin to the RET gene creates a novel rearranged form (PTC8) of the RET proto-oncogene in radiation-induced childhood papillary thyroid carcinoma. Cancer Res. 2000;60(11):2786–2789. [PubMed] [Google Scholar]
- 93.Zou M, et al. Concomitant RAS, RET/PTC, or BRAF mutations in advanced stage of papillary thyroid carcinoma. Thyroid. 2014;24(8):1256–1266. doi: 10.1089/thy.2013.0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Le Pennec S, et al. Intratumor heterogeneity and clonal evolution in an aggressive papillary thyroid cancer and matched metastases. Endocr Relat Cancer. 2015;22(2):205–216. doi: 10.1530/ERC-14-0351. [DOI] [PubMed] [Google Scholar]
- 95.Soares P, et al. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene. 2003;22(29):4578–4580. doi: 10.1038/sj.onc.1206706. [DOI] [PubMed] [Google Scholar]
- 96.Wojcicka A, et al. Variants in the ATM-CHEK2-BRCA1 axis determine genetic predisposition and clinical presentation of papillary thyroid carcinoma. Genes Chromosomes Cancer. 2014;53(6):516–523. doi: 10.1002/gcc.22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaczmarek-Rys, M., et al., The c.470 T > C CHEK2 missense variant increases the risk of differentiated thyroid carcinoma in the Great Poland population. Hered Cancer Clin Pract, 2015. 13(1): p. 8. [DOI] [PMC free article] [PubMed]
- 98.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gewinner C, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16(2):115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 101.Abubaker J, et al. Clinicopathological analysis of papillary thyroid cancer with PIK3CA alterations in a middle eastern population. J Clin Endocrinol Metab. 2008;93(2):611–618. doi: 10.1210/jc.2007-1717. [DOI] [PubMed] [Google Scholar]
- 102.Renaud F, et al. MUC1 expression in papillary thyroid carcinoma is associated with BRAF mutation and lymph node metastasis; the latter is the most important risk factor of relapse. Thyroid. 2014;24(9):1375–1384. doi: 10.1089/thy.2013.0594. [DOI] [PubMed] [Google Scholar]
- 103.Sheng H, et al. Prognostic significance of TMPRSS4 in gastric cancer. Neoplasma. 2014;61(2):213–217. doi: 10.4149/neo_2014_027. [DOI] [PubMed] [Google Scholar]
- 104.Jung H, et al. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene. 2008;27(18):2635–2647. doi: 10.1038/sj.onc.1210914. [DOI] [PubMed] [Google Scholar]
- 105.Larzabal L, et al. Overexpression of TMPRSS4 in non-small cell lung cancer is associated with poor prognosis in patients with squamous histology. Br J Cancer. 2011;105(10):1608–1614. doi: 10.1038/bjc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jarzab B, et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65(4):1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 107.Kebebew E, et al. ECM1 and TMPRSS4 are diagnostic markers of malignant thyroid neoplasms and improve the accuracy of fine needle aspiration biopsy. Ann Surg. 2005;242(3):353–361. doi: 10.1097/01.sla.0000179623.87329.6b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Forbes SA, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martin M, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karunamurthy A, et al. Prevalence and phenotypic correlations of EIF1AX mutations in thyroid nodules. Endocr Relat Cancer. 2016;23(4):295–301. doi: 10.1530/ERC-16-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mond M, et al. Somatic mutations of FOXE1 in papillary thyroid Cancer. Thyroid. 2015;25(8):904–910. doi: 10.1089/thy.2015.0030. [DOI] [PubMed] [Google Scholar]
- 112.Penna-Martinez M, et al. FOXE1 association with differentiated thyroid cancer and its progression. Thyroid. 2014;24(5):845–851. doi: 10.1089/thy.2013.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6(8):611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 114.Moyzis RK, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bae JS, et al. Clinical utility of TERT promoter mutations and ALK rearrangement in thyroid cancer patients with a high prevalence of the BRAF V600E mutation. Diagn Pathol. 2016;11:21. doi: 10.1186/s13000-016-0458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu X, et al. TERT promoter mutations and their association with BRAF V600E mutation and aggressive clinicopathological characteristics of thyroid cancer. J Clin Endocrinol Metab. 2014;99(6):E1130–6. doi: 10.1210/jc.2013-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu X, et al. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle. 2013;12(10):1637–1638. doi: 10.4161/cc.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23(3):R143–R155. doi: 10.1530/ERC-15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Melo M, et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab. 2014;99(5):E754–E765. doi: 10.1210/jc.2013-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salajegheh A, et al. Interactive role of miR-126 on VEGF-A and progression of papillary and undifferentiated thyroid carcinoma. Hum Pathol. 2016;51:75–85. doi: 10.1016/j.humpath.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 121.Yoruker EE, et al. MicroRNA expression profiles in papillary thyroid carcinoma, benign thyroid nodules and healthy controls. J Cancer. 2016;7(7):803–809. doi: 10.7150/jca.13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee JC, et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer. 2013;119(24):4358–4365. doi: 10.1002/cncr.28254. [DOI] [PubMed] [Google Scholar]
- 123.He H, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pallante P, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 125.Zhang JG, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411(11–12):846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 126.Lei ST, et al. MiR-639 promoted cell proliferation and cell cycle in human thyroid cancer by suppressing CDKN1A expression. Biomed Pharmacother. 2016;84:1834–1840. doi: 10.1016/j.biopha.2016.10.087. [DOI] [PubMed] [Google Scholar]
- 127.Hong S, et al. MiR-20b displays tumor-suppressor functions in papillary thyroid carcinoma by regulating the MAPK/ERK signaling pathway. Thyroid. 2016;26(12):1733–1743. doi: 10.1089/thy.2015.0578. [DOI] [PubMed] [Google Scholar]
- 128.Samsonov R, et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol. 2016;37(9):12011–12021. doi: 10.1007/s13277-016-5065-3. [DOI] [PubMed] [Google Scholar]
- 129.Hu J, et al. Expressions of miRNAs in papillary thyroid carcinoma and their associations with the clinical characteristics of PTC. Cancer Biomark. 2017;18(1):87–94. doi: 10.3233/CBM-161723. [DOI] [PubMed] [Google Scholar]
- 130.Thyroid Cancer: Follicular Cancer The Second Most Common Type of Thyroid Cancer. Available from: http://www.endocrineweb.com/conditions/thyroid-cancer/thyroid-cancer-follicular-cancer.
- 131.Sun D, et al. Microrna-199a-5p functions as a tumor suppressor via suppressing connective tissue growth factor (CTGF) in follicular thyroid carcinoma. Med Sci Monit. 2016;22:1210–1217. doi: 10.12659/MSM.895788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Y, et al. Genomic binding of PAX8-PPARG fusion protein regulates cancer-related pathways and alters the immune landscape of thyroid cancer. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smallridge RC, et al. American Thyroid Association guidelines for management of patients with anaplastic thyroid cancer. Thyroid. 2012;22(11):1104–1139. doi: 10.1089/thy.2012.0302. [DOI] [PubMed] [Google Scholar]
- 134.Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7(10):569–580. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- 135.Ricarte-Filho JC, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009;69(11):4885–4893. doi: 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kebebew E, et al. Anaplastic thyroid carcinoma. Treatment outcome and prognostic factors. Cancer. 2005;103(7):1330–1335. doi: 10.1002/cncr.20936. [DOI] [PubMed] [Google Scholar]
- 137.Landa I, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126(3):1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guan H, et al. Transmembrane protease serine 4 promotes thyroid cancer proliferation via CREB phosphorylation. Thyroid. 2015;25(1):85–94. doi: 10.1089/thy.2014.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Santarpia L, et al. Phosphatidylinositol 3-kinase/akt and ras/raf-mitogen-activated protein kinase pathway mutations in anaplastic thyroid cancer. J Clin Endocrinol Metab. 2008;93(1):278–284. doi: 10.1210/jc.2007-1076. [DOI] [PubMed] [Google Scholar]
- 140.Guerra, A., et al., Genetic mutations in the treatment of anaplastic thyroid cancer: a systematic review. BMC Surg, 2013. 13 Suppl 2: p. S44. [DOI] [PMC free article] [PubMed]
- 141.Kasaian K, et al. The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer. 2015;15:984. doi: 10.1186/s12885-015-1955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kunstman JW, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015;24(8):2318–2329. doi: 10.1093/hmg/ddu749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bonhomme B, et al. Molecular pathology of anaplastic thyroid carcinomas: a retrospective study of 144 cases. Thyroid. 2017; [DOI] [PubMed]
- 144.Latteyer S, et al. Targeted next-generation sequencing for TP53, RAS, BRAF, ALK and NF1 mutations in anaplastic thyroid cancer. Endocrine. 2016;54(3):733–741. doi: 10.1007/s12020-016-1080-9. [DOI] [PubMed] [Google Scholar]
- 145.Zhang WL, et al. miR-206 inhibits metastasis-relevant traits by degrading MRTF-A in anaplastic thyroid cancer. Int J Oncol. 2015;47(1):133–142. doi: 10.3892/ijo.2015.2993. [DOI] [PubMed] [Google Scholar]
- 146.Pagan, M., et al., The diagnostic application of RNA sequencing in patients with thyroid cancer: an analysis of 851 variants and 133 fusions in 524 genes. BMC Bioinformatics, 2016. 17 Suppl 1: p. 6. [DOI] [PMC free article] [PubMed]
- 147.Hu S, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119(10):2322–2329. doi: 10.1002/ijc.22110. [DOI] [PubMed] [Google Scholar]
- 148.Wang D, et al. RUNX3 site-specific hypermethylation predicts papillary thyroid cancer recurrence. Am J Cancer Res. 2014;4(6):725–737. [PMC free article] [PubMed] [Google Scholar]
- 149.Rodriguez-Rodero S, et al. DNA methylation signatures identify biologically distinct thyroid cancer subtypes. J Clin Endocrinol Metab. 2013;98(7):2811–2821. doi: 10.1210/jc.2012-3566. [DOI] [PubMed] [Google Scholar]
- 150.Mancikova V, et al. DNA methylation profiling of well-differentiated thyroid cancer uncovers markers of recurrence free survival. Int J Cancer. 2014;135(3):598–610. doi: 10.1002/ijc.28703. [DOI] [PubMed] [Google Scholar]
- 151.Bisarro Dos Reis M, et al., Prognostic Classifier Based on Genome-Wide DNA Methylation Profiling in Well-Differentiated Thyroid Tumors. J Clin Endocrinol Metab, 2017. 102(11): p. 4089–4099. [DOI] [PMC free article] [PubMed]
- 152.Beltrami CM, et al. Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas. Clin Epigenetics. 2017;9:45. doi: 10.1186/s13148-017-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]