Figure 5.
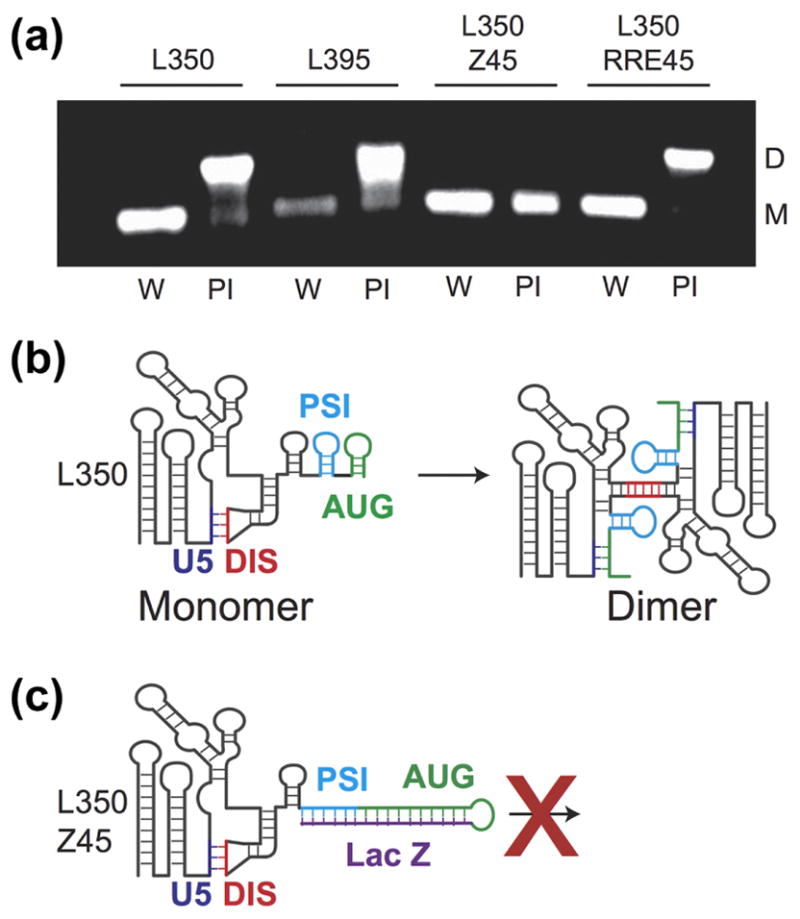
Influence of residues downstream of the 5′ UTR on the monomer-dimer RNA equilibrium. (A) Each construct was incubated overnight in water only (W) to form a monomer (M) or in physiological-like ionic strength (PI) buffer. L350 and L395 form a dimer (D) in PI buffer as expected. Addition of the non-native LacZ residues prevents dimerization in the L350-Z45 construct. L350-RRE retains dimerization behavior similar to the L350 and L395. (B) Cartoon representation of the 5′ UTR structure with the additional residues. In the monomer conformation, the region containing the AUG start codon forms a hairpin (green) and the dimer-promoting DIS hairpin (red) is sequestered by an intramolecular interaction with the U5 (blue). In the dimer conformation long-range base-pairing interactions between the AUG and U5 help expose the DIS hairpin and its palindromic sequence [39]. (C) The non-native residues in the L350-Z45 construct form a highly stable extended hairpin with AUG preventing the U5:AUG interaction, exposure of the DIS hairpin and thus formation of the dimer.
