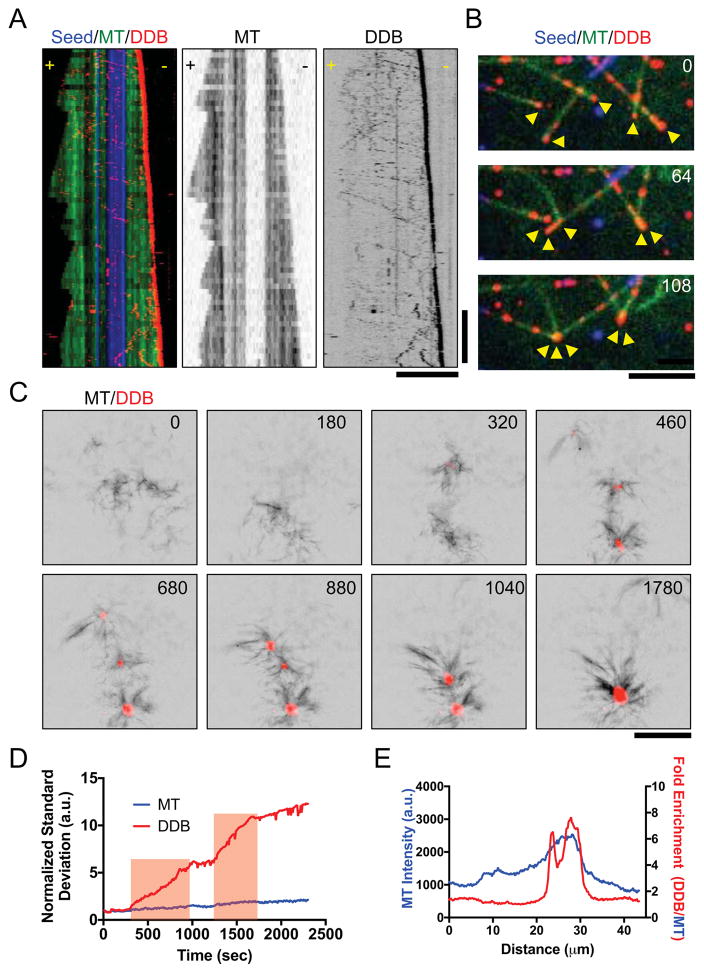
Figure 4. Minus-End Accumulations of DDB Drive Mesoscale MT Reorganization.
(A) Three-color kymograph of processive DDB (red) reveals strong accumulation at the growing minus-ends of dynamic MTs (green). Dynamic MT ends grown from GMPCPP-stabilized seeds (blue). Right: Individual fluorescent channels are reproduced for dynamic MTs and DDB for clarity. Scale Bars: 5 μm and 2 minutes. (B) Example of DDB accumulations (red) on growing MT minus-ends (green) driving MT-MT sliding to form minus-end clusters (arrows). Note that MT-MT sliding does not occur at MT crossover points not in contact with minus-end DDB clusters. Time in sec. Scale Bar: 5 μm. (C) Images of DDB-driven reorganization of growing microtubules (black) into asters with DDB (red) centers. Multiple asters coalesce into a single larger structure. Scale bar: 10 μm. Time in sec. (D) Plot of intensity contrast of both MT (blue) and DDB (red) channels expressed as standard deviation over time. Red bars indicate periods of merging between two asters. (E) Line scan plot of MT intensity (blue) across an aster and the fold increase of DDB fluorescence intensity divided by MT fluorescence intensity.