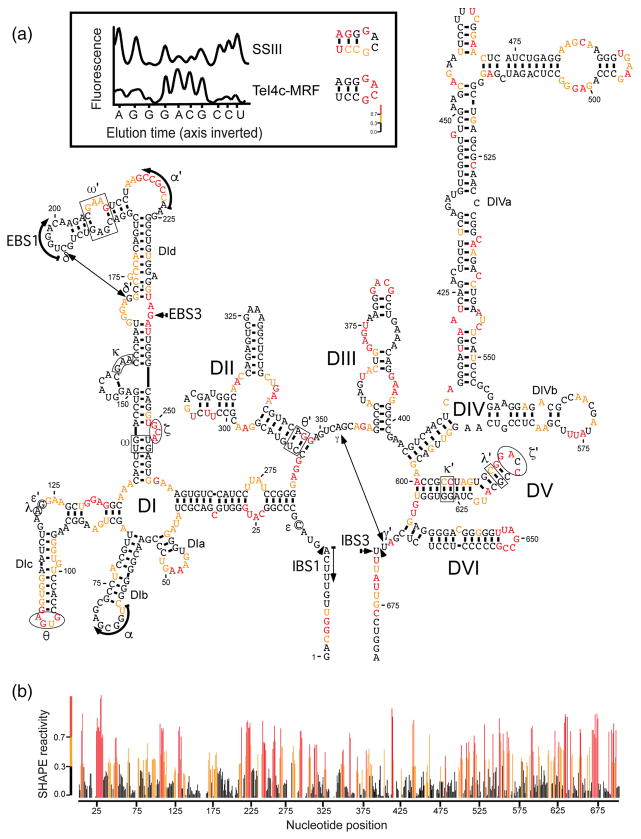
Fig. 2.
SHAPE analysis of GsI-IIC. SHAPE was done on an in vitro transcript containing the GsI-IIC-ΔORF+ΔA intron and short flanking exons with isatoic anhydride in reaction medium containing 5 mM Mg2+, as described in Materials and Methods. Modification sites were analyzed by primer extension with TeI4c-MRF RT [37] at 60 °C using a fluorescently labeled primer an-nealed near the 3′ end of the RNA, followed by capillary electrophoresis of the resulting cDNAs. (a) Predicted secondary structure of the GsI-IIC-ΔORF+ΔA intron showing SHAPE reactivities. Nucleotides are numbered from the 5′ end of the RNA used in the experiment. Nucleotide sequences involved in long-range tertiary interactions are boxed, circled, or indicated by arrows and are named with Greek letters. Colors indicate levels of SHAPE reactivities according to the scale in panel b below. The insert at the top compares cDNA raw traces produced by TeI4c-MRF and SuperScript III (SSIII) RTs. Peaks in the raw trace from capillary electrophoresis represent reverse transcription stops at single nucleotide resolution. Premature stops during primer extensions with SSIII RT created false-positive SHAPE reactivities, whereas TeI4c-MRF produced results expected for a group IIC intron. (b) Plot of SHAPE reactivity across the GsI-IIC intron. SHAPE reactivities were calculated for each nucleotide by using QuSHAPE [55]. SHAPE reactivities: red, high; orange, medium; black, none.