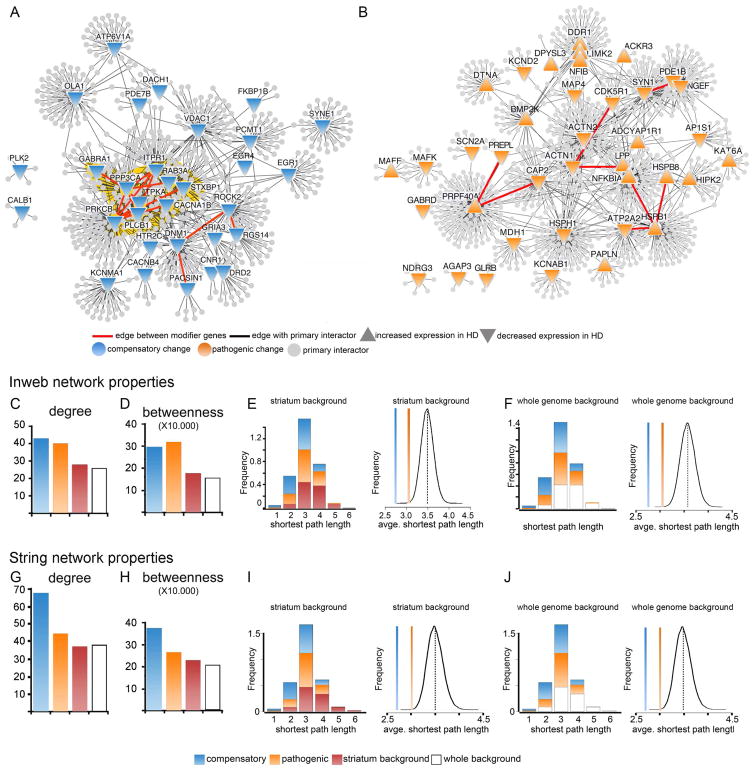
Figure 2. Network analysis of the compensatory and pathogenic subnetworks.
(A and B) Subnetworks of the potentially compensatory (A) and pathogenic (B) genes from the HD transcriptome showing their primary interactors. (C) Average node degree (number of edges per node) among the compensatory and pathogenic genes compared to the striatum (Wilcoxon rank sum test, p=5.41e-4 and 6.85e-3, respectively) and whole genome (p=1.38e-4 and 1.81e-3, respectively) backgrounds indicates that the compensatory and pathogenic modifiers are more tightly connected than randomly expected. This is also supported in (D) by the average betweenness (number of shortest paths crossing each node) of compensatory and pathogenic genes compared to striatum (p= 7.58e-5 and 3.33e-4, respectively) or whole genome (p= 1.79e-5 and 7.26e-5, respectively). (E-F) Network properties also show a higher connectivity among the genes in the compensatory and pathogenic subnetworks than would be randomly expected. (E) Shortest path length distribution among the compensatory and pathogenic genes compared to the striatum background and average shortest path length within the compensatory and pathogenic subnetworks compared to the striatal background (p=0 and 9.6e-4 respectively). P calculated by performing a background probability distribution (black line) corresponding to the 1.0e5 randomized samplings we ran using the striatal background genes. (F) Same as E but compared to the whole genome background (p= 0 and 1.3e-4 respectively). P calculated by performing a background probability distribution (black line) corresponding to the 1.0e5 randomized samplings we ran using the striatal background genes from the Inweb network. (G–J) Network properties of compensatory and pathogenic genes using the STRING network. Compensatory changes show the same behavior as with Inweb. Betweenness centrality is significantly increased (Wilcoxon rank sum p= 2.19e-6 for striatum and p= 4.72e-6 for whole genome). Node degree is significantly increased compared to striatum (Wilcoxon rank sum p= 6.42e-6) and whole genome (p= 3.37e-6) and average shortest path is significantly decreased (p=0). For the pathogenic subnetwork the average shortest path is significantly decreased (p=2.6e-4 striatum and 3e-5 whole genome). The betweenness centrality and degree indicators also show a trend to increase, but the Wilcoxon rank sum test does not return statistical significance.