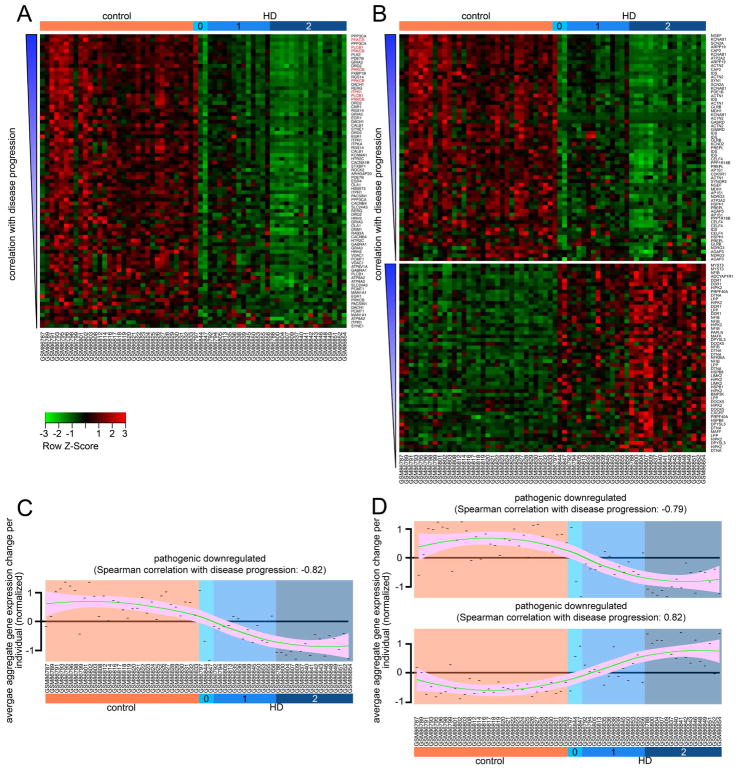
Figure 3. Reanalysis of human microarray data for compensatory and pathogenic changes and disease correlation analysis.
(A) Heat map of the gene expression changes in the compensatory network. Gene probes in each panel are organized by increasing degree of correlation with disease stage from bottom to top. Disease stage is indicated at the top, donor identifier at the bottom, and the gene symbol targeted by each probe on the right side. Genes in red font belong to the PLC cascade (PRKCB, PLCB1 and ITPR1). (B) Heat map of the GEP changes in the pathogenic network. Data arranged as in (A). (C and D) Trend lines representing the correlation between aggregate gene expression changes (per individual) with HD stages for each of the panels shown in A and B respectively. Note the tendency of the transcriptomic changes to become more pronounced in each group as the HD grade becomes more severe. Green line represents loess regression and pink shade indicates confidence interval. Each point is the average transcriptomic change for all the probes (in each group, indicated in the title) corresponding to a specific donor (indicated in the x axis). Chart in C corresponds to panel in A, and the same correspondence exists between charts in D and panels B. A–D (reanalyzed after (Hodges et al., 2006)). Correlation of gene expression changes with disease progression was calculated using Spearman’s correlation.