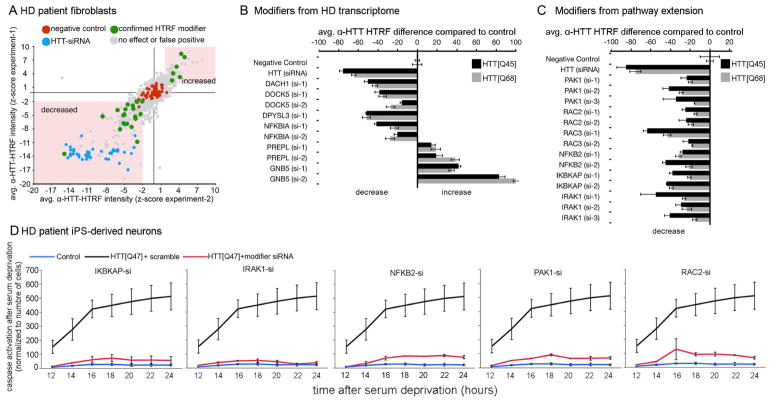
Figure 5. Knockdown of genes involved in inflammation/ regulation of actin cytoskeleton decreases mHTT protein levels in HD fibroblasts; pathway validation in HD iPS-derived neurons.
(A–B) Analysis of mHTT protein levels in fibroblasts from HD patients. Fibroblasts were transfected with siRNAs targeting the human homologs of the Drosophila genetic modifiers identified among genes altered in the HD transcriptome. (A) Scatter plot summarizing the HTRF screen in HTT[Q68] fibroblasts for genes modulating mHTT protein levels. All 82 human genes whose Drosophila homologs modified motor impairments caused by HTTN231Q128-induced neuronal dysfunction were targeted using 8 siR-NAs per gene. Screen was done in duplicate (experiment-1 and -2). (B) Effect of the hit genes on mHTT levels in HTT[Q68] and HTT[Q45] patient fibroblast lines normalized to negative control. Error bars: standard deviations. (C) Analysis of mHTT protein levels in HD fibroblasts transfected with siRNAs targeting the additional genetic modifiers identified by pathway extension analysis. Data is shown for both HTT[Q68] and HTT[Q45] fibroblast lines. (D) Charts showing average caspase-3 activation following BDNF deprivation as a function of time. Black line: iPS-derived neuron like cells from a HTT[Q47] patient. Red line: HTT[Q47]-derived neurons transfected with siRNAs targeting the indicated gene. Blue line: control iPS-derived neuron like cells. The donor was the sibling of the patient that donated the HTT[Q47] cells. Error bars indicate standard deviation. All the differences shown in B and C where significantly different compared to the corresponding negative controls (using Anova followed by Student’s t test, α=0.05).