FIGURE 2.
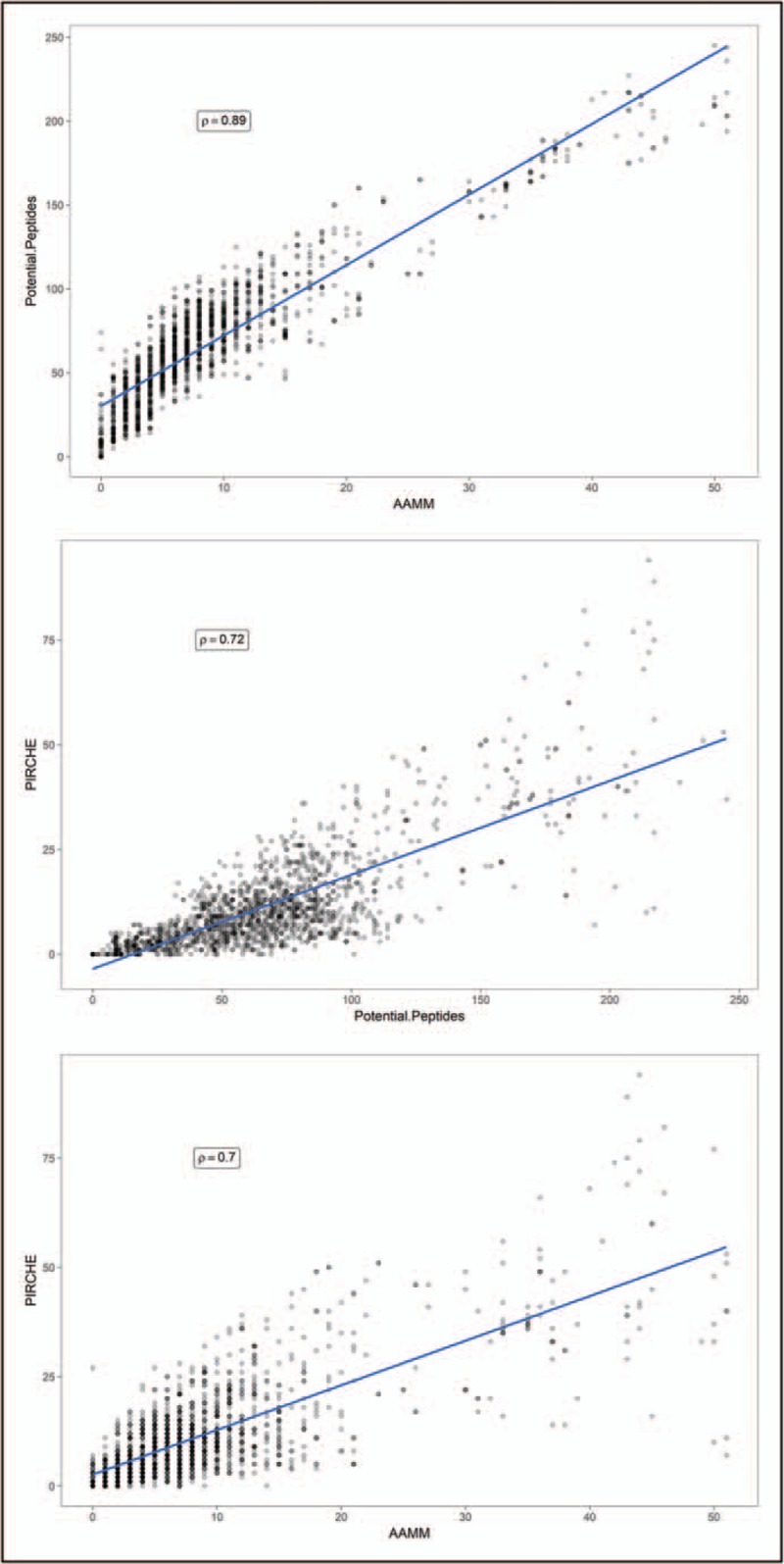
Correlation between predicted indirectly recognizable HLA epitopes and amino acid sequence polymorphism. The figure depicts analysis of HLA (A, B, C, DR, DQ and DP) mismatches in a local cohort of 182 donor–recipient pairs (HLA typed at two-field resolution). HLA mismatches were identified (n = 1615) and scored according to the number of possible peptide epitopes (unique, polymorphic nonameric binding cores) that can be derived from donor HLA (irrespective of predicted binding affinity to recipient HLA-DR), the number of predicted indirectly recognizable HLA epitopes (predicted indirectly recognizable HLA epitopes; defined as polymorphic nonameric binding cores with predicted binding to recipient HLA-DR of IC50 < 1000 nmol/l), and the number of amino acid mismatches (as defined by the Cambridge HLA immunogenicity algorithm). Donor HLA-derived polymorphic nonameric binding cores were defined as those that differed by at least one amino acid compared with the HLA sequences of the recipient. The NetMHCIIpan version 3.1 algorithm was implemented locally to calculate the predicted indirectly recognizable HLA epitopes scores (the predicted indirectly recognizable HLA epitopes algorithm available via https://www.pirche.org implements a previous version of NetMHCIIpan). For the purposes of this analysis, all HLA mismatches were grouped together. There was high correlation between scores (Spearman's Rho), although this correlation varied for mismatches within individual HLA loci (data not shown). HLA, human leukocyte antigen.
