Extended Data Figure 5. The histidine degradation pathway affects the sensitivity of cancer cells to methotrexate and HAL expression is associated with treatment response in acute lymphoblastic leukemia (ALL) patients (part 2).
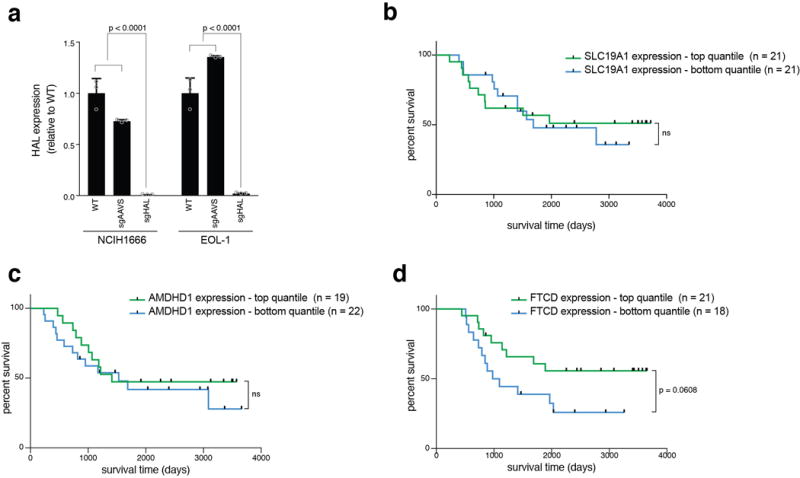
a. Validation of genetic depletion of HAL by the CRISPR/Cas9 system in NCIH1666 and EOL-1 cells. Expression levels of HAL were measured by qPCR and normalized to the average of two control genes, UBC and HPRT. p-values were calculated by two-way ANOVA. n=3 technical replicates. Error bars indicate SD. Raw data for survival assays Fig. 3e, f can be found in Source Data_3, sheets “cell lines survival” and “HAL KO survival”. b-d. Expression levels of SLC19A1 (b), AMDHD1 (c), or FTCD (d) do not predict survival of pediatric ALL patients treated with a regimen that includes methotrexate. Kaplan-Meier curves of overall survival of ALL patients with high (top quantile, colored green) and low (bottom quantile, colored blue)24 expression of each of the assayed genes. Patient sample size for each group is indicated. p-values were calculated using the log-rank (Mantel-Cox) test. Raw data for ALL patients gene expression can be found in Source Data_3, sheet “ALL patients”.
