Figure 4. CCR2- and CCR2+ macrophages display distinct gene expression profiles.
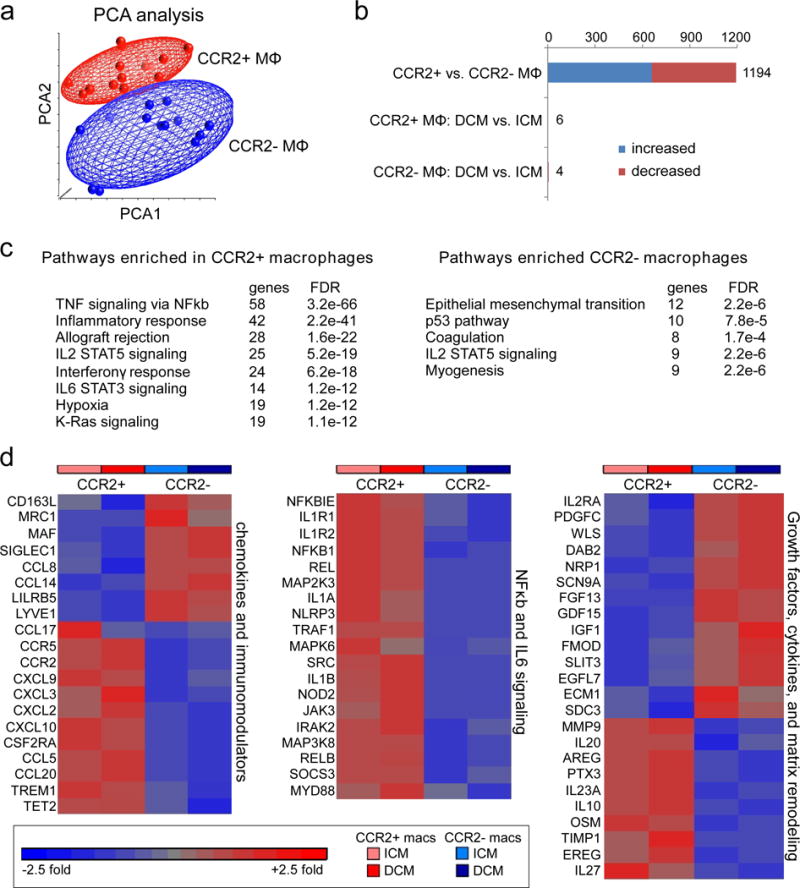
a, Principal component analysis of CCR2- (n=19) and CCR2+ (n=19) cardiac macrophages (ICM and DCM). Red: CCR2+ macrophages, Blue: CCR2- macrophages. b, Bar graph displaying the number of genes that were differentially expressed between all CCR2- and CCR2+ macrophages (DCM and ICM) and the number of genes that were differentially expressed in CCR2- and CCR2+ macrophages stratified by DCM and ICM designation using a threshold of 1.4X fold change and FDR<0.05. Blue: increased expression, Red: decreased expression. c, GSEA pathway analysis revealing pathways enriched in cardiac CCR2+ versus CCR2- macrophages (ICM and DCM). Statistical significance was evaluated using false discovery rate (FDR). d, Heat maps showing relative fold changes in genes associated with chemokine and immunomodulatory signaling, NFκb and IL6 signaling, as well as selected growth factors, cytokines, and extracellular matrix remodeling factors. Data are shown for CCR2+ and CCR2- macrophages obtained from specimens of patients with ICM or DCM and the results are displayed as average expression values. All genes displayed on the heat maps were differentially expressed (FDR<0.05) between CCR2+ and CCR2- macrophages.
