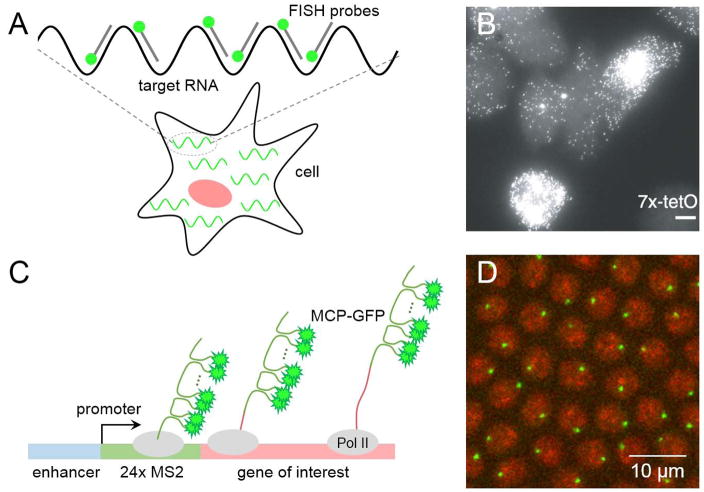
Figure 2. Imaging techniques for visualizing nascent transcription activity.
(A) Schematic of single molecule fluorescent in situ hybridization (smFISH). Several short oligo probes that are complementary to the target RNA are designed to detect individual RNA molecules. Binding of multiple fluorescently labeled probes to the target RNA provides enough sensitivity and allows detection of single nascent RNA transcripts within cells.
(B) A representative image of cells from clonal cell lines where each mRNA is hybridized to smFISH probes (image from [6]).
(C) Schematic of MS2-MCP imaging method. The system uses bacteriophage MS2 and MS2 coat proteins (MCP) that is labeled with fluorescent proteins such as GFP. Typically, 6–24 tandem repeats of MS2 sequences are being inserted to the 5′ or 3′ untranslated region (UTR) of the gene of interest. Upon transcription, MS2 forms a stem loop and dimeric MCP-GFP binds to each stem loop, allowing the detection of de novo transcripts through fluorescent imaging.
(D) A representative snapshot of an early Drosophila embryo, where nascent transcripts are visualized with the MS2-MCP system. Nuclei are shown in red and nascent transcripts are shown in green.