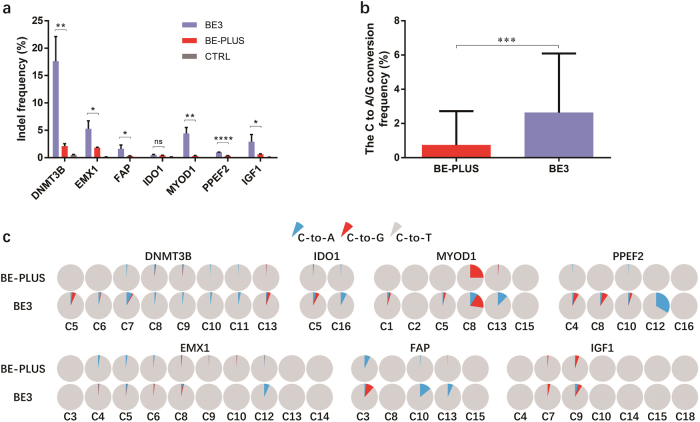
Fig. 3.
BE-PLUS significantly enhances the fidelity of base editing. a Analysis of indels by deep sequencing. A total of 69,100,000 reads were generated and analysed. Reads containing at least 1 inserted or deleted nucleotides ±20 bp surrounding the protospacers were calculated as indel-containing reads. Indel frequency was calculated as the number of indel-containing reads among the total number of mapped reads. CTRL, negative control. Error bars (±) indicate the standard deviations of 3 replicates. ns (not significant), P ≥ 0.05; *P < 0.05; **P < 0.01; ****P < 0.000 1. b Analysis of unwanted base conversions. The average frequencies of C-to-A and C-to-G conversions of all mutant products induced by BE-PLUS and BE3 are presented. Error bars (±) indicate the standard deviations. ***P < 0.001. c Individual fractions of C-to-T, C-to-A and C-to-G conversions at 7 loci induced by BE-PLUS and BE3. The data shows a representative experiment from three independent experiments